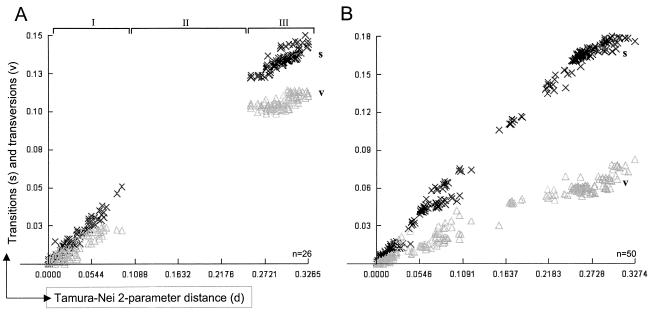
FIG. 2.
Transitions and transversions of PERV sequences plotted versus Timura-Nei′s two-parameter genetic distance. Nucleotide sequences of LTRs (A) and env (B) used to determine the evolutionary age of PERV were analyzed with the program DAMBE (44) for their transitions (× and s) and transversions (Δ and v) ratio employing the Tamura-Nei substitution model (36). Transitions are nucleotide changes T↔C or A↔G and should happen more frequently than transversions, which are changes encompassing T↔A, T↔G, C↔A, or C↔G. While the s/v plot of env sequences (B) shows a linear increase of both transitions and transversions, with transitions progressing much faster, the s/v plot of LTR sequences (A) shows a gap (II) based on LTRs with different numbers of repeats, leading to a stepwise increase in sequence variations. When comparing viruses with different LTR types, each repeat-harboring LTR type increases genetic distance by 39 bp, while the repeatless LTRs deviate considerably from the repeat-harboring LTRs on the basis of pairwise comparison, thus producing the gap in genetic distance that is observed in the LTR s/v plot. The significant dominance of transitions and no crossing of the different symbols, representing transitions and transversions, suggest no substitution saturation and proof of a constant rate of evolution, therefore allowing a determination of age by molecular clock. Each × or Δ in the plot represent a transition or transversion event at a given genetic distance, respectively. n, number of sequences analyzed for each plot (see Appendix).