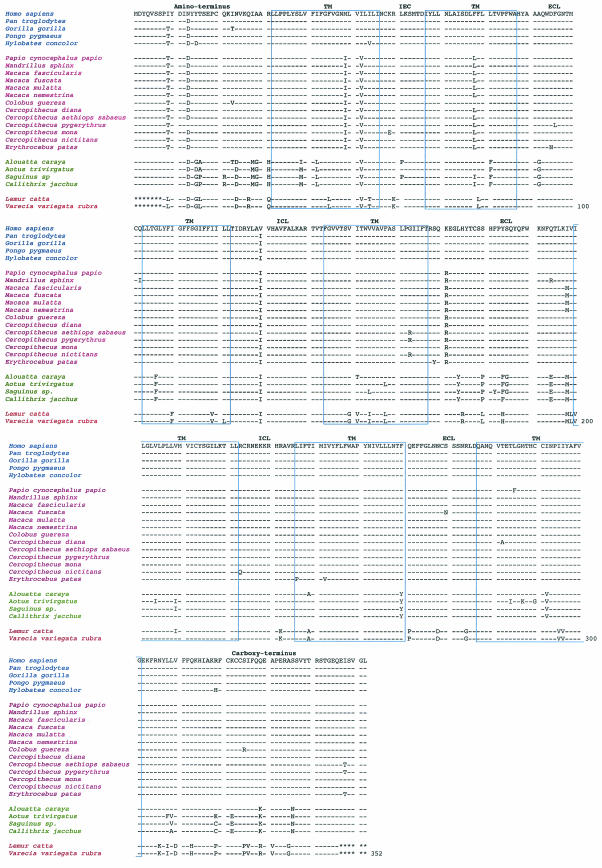
FIG. 2.
Alignment of deduced human and nonhuman primate CCR5 homologue amino acid sequences. Directly below the human CCR5 consensus sequence are the nonhuman primate CCR5 homologue sequences for apes (P. troglodytes, G. gorilla, P. pygmaeus, and H. concolor), Old World monkeys (P. cynocephalus papio, M. sphinx, M. fascicularis, M. fuscata, M. mulatta, M. nemestrina, C. guereza, C. diana, C. aethiops sabaeus, C. pygerythrus, C. mona, C. nictitans, and E. patas), New World monkeys (A. caraya, A. trivirgatus, Saguinus sp., and C. jacchus), and prosimians (L. catta and V. variegata rubra). Each nonhuman primate CCR5 homologue sequence is the consensus sequence of six clones obtained from each animal to ensure a 97% probability that both alleles were represented. Amino acids in the alignment that share similarity with the human CCR5 consensus sequence shown at the top are indicated by a dash. The truncated 5′ and 3′ ends of the prosimian CCR5 homologues are indicated by asterisks. The amino acid alignment is annotated for the amino terminus, extracellular loops (ECL), intracellular loops (ICL), transmembrane regions (TM), and carboxy terminus. Apes are highlighted in blue, Old World monkeys are highlighted in magenta, New World monkeys are highlighted in green, and prosimians are highlighted in red.