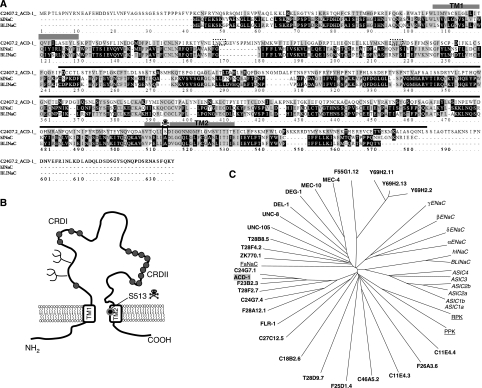
Figure 1.
The C. elegans DEG/ENaC channel ACD-1 (acid-sensitive channel degenerin-like) shares similarity with mammalian BLINaC and hINaC. (A) We designated C24G7.2 as ACD-1 because of its sensitivity to acidic solutions. C24G7.2 protein sequence and alignments with hINaC (29% identity and 45% similarity) and mouse and rat BLINaC (28% identity and 44% similarity). The transmembrane domains TM1 and TM2 and the region of the protein deleted in acd-1(bz90) knockout are indicated by the grey boxes and black line, respectively. The transmembrane domains were identified by threading ACD-1 sequence onto the sequence of chicken ASIC1a (Jasti et al, 2007), using PyMOL (www.PyMOL.org). The box and the skull designate position S513 in ACD-1 corresponding to the amino acid that when mutated to Val or Thr in MEC-4 induces hyperactivation of the channel (A713V or T, the (d) mutation). The dotted boxes indicate potential glycosylation sites. Alignment by ClustalW (available at bioweb.pasteur.fr). (B) A schematic diagram representing ACD-1 predicted topology. The potential glycosylation sites are indicated by the branches. The circles depict cysteine residues; a region just preceding TM2 is rich in cysteines. This feature is conserved in DEG/ENaC subunits across species (Benos and Stanton, 1999). (C) Dendrogram of C. elegans (bold), Drosophila (RPK and PPK, underlined)), Helix apersa (FaNaC, underlined) and mammalian (italic) DEG/ENaC channel subunits. ACD-1 subunit has a grey background.