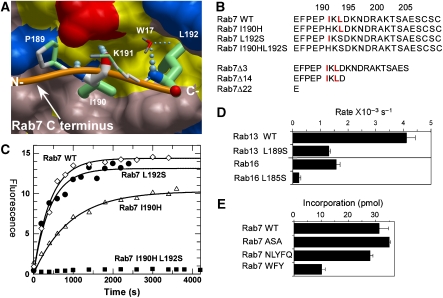
Figure 5.
Analysis of the CBR–CIM interaction. (A) Structural basis of the REP-1 CBR interaction with the CIM of Rab7. The hydrophobic CBR patch of REP-1 is displayed in surface representation, and hydrophobic residues are coloured yellow, and polar and charged residues are coloured in pink, blue (+), and red (−). The main chain atoms involved in hydrogen bonding are displayed in atomic colours. The C terminus of Rab7 is displayed as an orange worm, and the residues RabP189-RabL192 are displayed in stick format. N- and C- denote the termini. Atoms involved in hydrophobic interactions are coloured in green. The boldness of the stick representation is an indication of the degree of interaction with REP. The order of contact area for these residues is L192>I190>P189>K191. W17 denotes a water molecule involved in the hydrogen bonding network. (B) C-terminal mutants of Rab7 used in this study. The CIM motif is highlighted in red. Mutation of I190 to histidine was chosen based on the alignment with Rab27A, which does not display a canonical CIM motif. (C) In vitro prenylation of wild-type Rab7 and its CIM mutants with NBD-FPP. The amount of prenylated substrate was determined by fluorescent scanning of SDS–PAGE-resolved samples. (D) Changes in in vitro prenylation rates of wild-type and CIM mutants of Rab13 and Rab16. (E) Effect of extension of the Rab7 C terminus on the ability to serve as substrates of RabGGTase.