Abstract
Sequence analysis of the penA gene, encoding penicillin-binding protein 2 (PBP2), in 30 penicillin-intermediate (PenI) Neisseria meningitidis strains showed altered gene sequences due to the translocation of exogenous DNA blocks derived from commensal neisseriae, which are known to have PBP2 proteins with decreased affinity for the antibiotic. In order to obtain a rapid and reproducible method for predicting the PenI phenotype, a real-time PCR assay was set up with primers and probes designed on the basis of the penA gene. The A→G mutation at codon 566, in the transpeptidase domain of the penA gene (which is present in the whole sample of 30 PenI strains and in all the 41 sequences of PenI meningococci isolated worldwide and has been deposited in the sequence databank), was chosen as a marker of penA translocations. Two hybridization probes were designed to distinguish the wild-type penA gene in penicillin-susceptible (PenS) meningococci from the mutated penA gene at codon 566 in PenI strains. Thermal analysis of probe hybridization revealed a melting temperature difference of at least 6°C between PenI and PenS strains. This real-time PCR protocol characterizes the penicillin phenotype of N. meningitidis in a few hours without DNA sequencing and is useful for rapid screening of the penicillin-intermediate genotype among meningococcal isolates.
Neisseria meningitidis, a human pathogen, is a leading cause of life-threatening infections, such as meningitis and sepsis. The treatment of meningococcal infections with penicillin G is still effective, but the number of reports describing the isolation of meningococci with decreased susceptibility to penicillin in different countries is on the rise (4, 6; P. Botha, Letter, Lancet i:54, 1988; A. P. Gomes and R. Siqueira-Batista, Letter, Rev. Assoc. Med. Bras. 48:114, 2002; J. A. Saez-Nieto, J. A. Vazquez, and C. Marcos, Letter, Lancet 336:54, 1990). This phenomenon is caused by the emergence of penA genes whose sequences are altered, determining the production of penicillin-binding proteins (PBPs) with a lower affinity for β-lactam antibiotics. The sequences of these modified penA genes are highly variable. As described previously (1), these alterations are due to genetic exchange, in the human nasopharynx, between N. meningitidis and naturally resident nonpathogenic neisseriae, resulting in a mosaic structure of the target gene. penA sequences from penicillin-intermediate (PenI) strains have been extensively analyzed in different studies (1-3, 12, 13), particularly in the transpeptidase-encoding region (nucleotides 571 to 1746), in order to characterize the genetic polymorphism and to demonstrate a correlation with reduced susceptibility to penicillin. The alteration of only one PBP (PBP2) in N. meningitidis suggests that strains with decreased susceptibility are still evolving, so the emergence of penicillin-resistant strains may be expected in the near future, as has been observed for Streptococcus pneumoniae (12). The development of molecular methods to monitor the changes in the penA genes of meningococci is hence warranted.
This study proposes a reliable and rapid molecular approach to the detection of reduced susceptibility to penicillin in invasive N. meningitidis strains. In the absence of a single specific determinant of decreased susceptibility to penicillin in N. meningitidis, a real-time PCR assay has been developed for the identification of nucleotide substitutions within the transpeptidase domain of the penA gene by oligonucleotide hybridization and thermal analysis. This method permits rapid and reproducible screening of the amino acid substitution Ile566→Val (ATT to GTT). In fact, this mutation has been detected in all 30 PenI strains isolated in Italy and in 41 penA sequences of PenI meningococci isolated worldwide and deposited in the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov); it has never been detected in penicillin-susceptible (PenS) strains.
MATERIALS AND METHODS
Bacterial strains and penicillin susceptibility test.
Five hundred sixteen strains of N. meningitidis isolated from patients were received by the reference laboratory at the Istituto Superiore di Sanitá during the period 1995 to 2002. MICs were determined by E-test (AB Biodisk) on Mueller-Hinton agar (Oxoid) supplemented with 5% laked horse blood and incubated in air with 5% CO2 at 35°C for 24 h. The breakpoints were those recommended for Neisseria gonorrhoeae by the National Committee for Clinical Laboratory Standards: for susceptibility, ≤0.06 μg/ml; for resistance, ≥2 μg/ml (8).
For the real-time PCR assay, a panel consisting of 30 PenI strains isolated in different years and 36 PenS meningococci were examined. N. meningitidis serogroup B MC58 was used as a susceptible reference strain (Table 1).
TABLE 1.
Characteristics and Tm of 30 penicillin-intermediate and 36 penicillin-sensitive N. meningitidis strains
| Strain code (accession no. for penA sequence) | N. meningitidis PenI or PenS phenotype | Yr of isolation | MIC for penicillin (μg/ml) | Tm (°C) |
|---|---|---|---|---|
| PenI strains | ||||
| 661 (AJ549315) | C:2b:P1.2 | 1995 | 0.125 | 55.0 |
| 759 | C:2b:P1.2 | 1996 | 0.125 | 55.0 |
| 864 (AJ549316) | B:4:P1.12 | 1997 | 0.094 | 51.7 |
| 868 (AJ549309) | B:1:P1.3 | 1997 | 0.125 | 53.5 |
| 884 | C:2b:P1.5 | 1998 | 0.190 | 55.0 |
| 918 (AJ549310) | B:4:P1.16 | 1999 | 0.125 | 53.5 |
| 949 (AJ549311) | B:1:P1.15, 16 | 1999 | 0.094 | 53.5 |
| 963 (AJ549312) | B:4, 15:P1.16 | 1999 | 0.125 | 53.5 |
| 990 (AJ549313) | C:nt:P1.16 | 1999 | 0.094 | 53.5 |
| 1000 (AJ549314) | B:14:P1.13 | 1999 | 0.190 | 53.5 |
| 1047 (AJ549307) | B:15:nst | 2000 | 0.125 | 53.5 |
| 1067 (AJ549308) | B:15:P1.7, 16 | 2000 | 0.190 | 53.5 |
| 1151 | C:2b:P1.5 | 2001 | 0.125 | 53.5 |
| 1173 | W135:nt:nst | 2001 | 0.094 | 55.0 |
| 1194 (AJ549305) | B:4:P1.4 | 2002 | 0.125 | 53.5 |
| 1208 | C:2b:P1.5 | 2002 | 0.190 | 55.0 |
| 1237 | C:2b:P1.5 | 2002 | 0.125 | 55.0 |
| 1230 (AJ549306) | B:nt:nst | 2002 | 0.094 | 53.5 |
| 1233 (AJ549302) | C:2a:P1.5 | 2002 | 0.250 | 53.5 |
| 1234 | C:2a:P1.5 | 2002 | 0.250 | 53.5 |
| 1228 (AJ549303) | C:2b:P1.5 | 2002 | 0.125 | 55.0 |
| 1235 | C:2b:P1.5 | 2002 | 0.125 | 55.0 |
| 1241 | C:2a:P1.5 | 2002 | 0.190 | 53.5 |
| 1245 | B:15:P1.16 | 2002 | 0.250 | 53.5 |
| 1258 | C:2b:P1.5 | 2002 | 0.250 | 55.0 |
| 1263 | C:nt:P1.5 | 2002 | 0.125 | 55.0 |
| 1271 (AJ549304) | B:1:P1.6 | 2002 | 0.094 | 53.5 |
| 1277 | B:nt:P1.16 | 2002 | 0.250 | 53.5 |
| 1284 | C:2b:P1.5 | 2002 | 0.190 | 55.0 |
| 1285 | C:2b:P1.5 | 2002 | 0.125 | 55.0 |
| PenS strains | ||||
| 659 | Poli:15:P1.7, 16 | 1994 | 0.047 | 45.5 |
| 690 | B:2b:P1.10 | 1995 | 0.032 | 45.5 |
| 898 | B:4, 15:P1.10, 16 | 1998 | 0.047 | 45.5 |
| 904 | B:21:P1.13 | 1998 | 0.012 | 45.5 |
| 921 | B:14:P1.13 | 1999 | 0.047 | 45.5 |
| 926 | B:14:P1.13 | 1999 | 0.047 | 45.5 |
| 927 | B:14:P1.13 | 1999 | 0.047 | 45.5 |
| 939 | B:4:P1.4 | 1999 | 0.047 | 45.5 |
| 1101 | B:15:P1.7, 16 | 2000 | 0.032 | 45.5 |
| 1109 | B:14:P1.13 | 2000 | 0.012 | 45.5 |
| 1120 | B:15:P1.4 | 2001 | 0.016 | 45.5 |
| 1122 | B:14:P1.13 | 2001 | 0.008 | 45.5 |
| 1129 | B:14:P1.13 | 2001 | 0.012 | 45.5 |
| 1130 | C:2b:P1.5 | 2001 | 0.032 | 45.5 |
| 1133 | C:2b:P1.5 | 2001 | 0.023 | 45.5 |
| 1135 | B:14:P1.13 | 2001 | 0.008 | 45.5 |
| 1136 | C:2b:P1.5 | 2001 | 0.023 | 45.5 |
| 1138 | B:nt:P1.4 | 2001 | 0.006 | 45.5 |
| 1140 | C:2b:P1.5 | 2001 | 0.032 | 45.5 |
| 1142 | W135:2a:P1.5, 2 | 2001 | 0.008 | 45.5 |
| 1147 | B:14:P1.13 | 2001 | 0.008 | 45.5 |
| 1150 | B:14:P1.13 | 2001 | 0.008 | 45.5 |
| 1157 | B:nt:nst | 2001 | 0.016 | 45.5 |
| 1159 | B:15:P1.7, 16 | 2001 | 0.008 | 45.5 |
| 1161 | B:15:P1.4 | 2001 | 0.012 | 45.5 |
| 1175 | B:14:P1.13 | 2001 | 0.012 | 45.5 |
| 1184 | B:14:P1.13 | 2001 | 0.080 | 45.5 |
| 1190 | W135:nt:P1.3 | 2001 | 0.064 | 45.5 |
| 1196 | C:2b:P1.5 | 2002 | 0.023 | 45.5 |
| 1198 | C:2a:P1.5 | 2002 | 0.064 | 45.5 |
| 1199 | C:2b:P1.5 | 2002 | 0.047 | 45.5 |
| 1203 | B:2a:P1.5, 2 | 2002 | 0.023 | 45.5 |
| 1221 | C:2b:P1.5 | 2002 | 0.047 | 45.5 |
| 1225 | C:2b:P1.5 | 2002 | 0.064 | 45.5 |
| 1254 | B:15:P1.4 | 2002 | 0.016 | 45.5 |
| 1267 | B:4:nst | 2002 | 0.047 | 45.5 |
DNA isolation, gene amplification, sequencing, and analysis.
Chromosomal DNA was extracted by using the QIAamp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
The coding regions of the penA genes (from nucleotides 546 to 1746) of all the strains examined were amplified by using the primers and PCR conditions described by Zhang et al. (15). The PCR products were further purified by use of QIAquick purification columns (Qiagen) for sequencing by the Sanger method (11a) with the ABI Prism DNA sequencer 377 (Perkin-Elmer). The sequences were analyzed with the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
Real-time PCR and oligonucleotide thermal analysis.
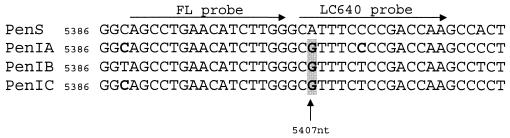
A primer-probe set was designed on the basis of the penA gene nucleotide mutations detected in PenI meningococci. The primers for the PCR amplification were primer1 (5′-GATTGTGGCGGTAACCATTGACG; nucleotide positions 5298 to 5320 in EMBL/GenBank accession no. AE002397) and primer2 (5′-CGGGGATATAACTGCGTCCGT; nucleotide positions 5644 to 5624 in accession no. AE002397 [14]), defining a region of about 350 bp. As shown in Fig. 1, the fluorescence probe (FL, donor dye) was designed at positions 5389 to 5404 (5′-AGCCTGAACATCTTGGFL-3′), in a region showing identical penA DNA sequences in PenS and PenI strains. The Red640 probe was positioned 1 bp away from the FL probe (5′-Red640CGTTTCTCCGACCAA-P-3′) and designed to be homologous to the sequences with accession no. AJ549302 to AJ549315.
FIG. 1.
LightCycler probe positions in a partial penA sequence alignment. Nucleotide mismatches in the PenI genotype (penIA, penIB, penIC) are boldfaced.
The real-time PCR mixture contained 1 μl of purified chromosomal DNA (100 ng), 1 μl (10 pmol) of each primer, 2 μl (2 pmol) of probes FL and LC640, 2.5 μl of MgCl2 (final concentration, 4 mM), 2 μl of Fast-start Master Hybridization Probes reaction mixture (Roche Diagnostic, Mannheim, Germany), and PCR-grade sterile water to a final volume of 20 μl. The samples were run by performing an initial 10-min denaturation step at 95°C followed by 40 cycles of repeated denaturation (10 s at 95°C), annealing (10 s at 48°C), polymerization (10 s at 72°C), and detection of the fluorescence (15 s at 38°C). The last step was added at the end of each PCR cycle to increase red fluorescence levels for the PCR quantitative analysis. The temperature transition rate was 20°C/s in all segments.
Fluorescence was measured in channel 2 at 640 nm. Data were analyzed with LightCycler software (version 5.32) according to the manufacturer's instructions (Roche Diagnostics).
In each run the susceptible reference strain, one PenI strain for which the penicillin MIC was defined, and a negative control (sterile water instead of a DNA template) were included for the reproducibility of results and to check for contamination.
Nucleotide sequence accession numbers.
Fifteen representative nucleotide sequences out of the 30 analyzed in the present study were deposited in the EMBL/GenBank nucleotide sequence databases under accession no. AJ549302 to AJ549316 (Table 1).
RESULTS
Susceptibility to penicillin and penA sequence analysis.
Strains with PenI phenotypes account for more than 10% of all meningococci isolated in Italy from 1995 to 2002. Interestingly, during the year 2002, a higher proportion of PenI meningococci was detected (26%), and the C:2b:P1.5 phenotype was predominant. Moreover, MICs for PenI strains were higher than those found in previous years (7). The phenotypic characteristics of all the strains examined are shown in Table 1.
The amino acid change from Ile566 to Val566 (ATT→GTT transition), found at the 3′ end of the penA gene in all the PenI meningococci examined, allowed us to design the probes used in the real-time PCR assay (Fig. 1). Four other nucleotide transitions (causing amino acid substitutions Phe504→Leu, Ala510→Val, Ile515→Val, and His541→Asn), derived from commensal neisseriae, were always associated with the Ile566→Val mutation. The mutation at codon 566 was chosen as a marker for penA translocation, since the other four mutations were localized within an extremely variable region that was not useful as a target sequence for primer hybridization and thermal analysis. The five mutations were also consistently detected in all 41 sequence alignments of PenI N. meningitidis strains deposited by other authors (http://www.ncbi.nlm.nih.gov).
LightCycler penA mutation assay.
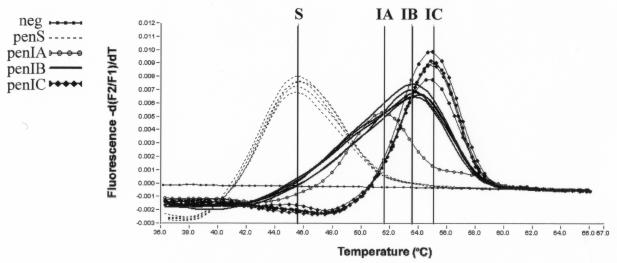
Oligonucleotide hybridization and thermal analysis, using the FL and LC640 probes, yielded different melting curves for susceptible N. meningitidis strains (45.5°C) and PenI strains carrying the ATT566→GTT mutation, which resulted in a melting temperature (Tm) increase of 6 to 9.5°C. Seventeen PenI strains that carried the mutation at codon 566 showed a Tm of 53.5°C (PenIB), whereas 12 PenI strains that also had a C→T transition showed a Tm of 55°C (PenIC) (Fig. 2). The presence of this nucleotide difference localized 1 base before the FL probe results in a 1.5°C increase in the thermal dissociation of the FL-LC640 probes. The fact that differences in neighbor bases can cause a slight change in the stability of the probes has been reported previously (10). One strain harboring the mutation at codon 566 also carried an additional nucleotide mismatch, a C instead of a T, compared to the LC640 probe sequence (Fig. 1) and showed a Tm corresponding to 51.7°C (PenIA) (Fig. 2). In summary, all PenI strains, showing the codon 566 mutation alone or in combination with other nucleotide substitutions, produced discernible peaks in the range from 51 to 55°C, allowing them to be distinguished from the PenS strains, which peaked at 45.5°C (Fig. 2). Results obtained by the real-time PCR assay were reproducible and always in agreement with the nucleotide sequencing data and MICs (Table 1). In fact, all PenI strains showed Tm values between 51.7 and 55°C, whereas all PenS strains showed a Tm of 45.5°C.
FIG. 2.
Tm curves for three different PenI genotypes (penIA, penIB, and penIC). All PenS strains peaked within the same curve. Melting peaks were derived by plotting the negative derivative of fluorescence with respect to temperature (−dF/dT).
DISCUSSION
Since the introduction of penicillin for the treatment of meningococcal infections, the occurrence of N. meningitidis strains resistant to penicillin due to β-lactamase-producing plasmids has been rare (4), whereas the frequency of meningococci with decreased susceptibility to the antibiotic has steadily increased (5, 11; Saez-Nieto et al., letter). This behavior is due to the well known lower penicillin affinity of the PBP2 encoded by DNA originating from nonpathogenic commensal nasopharyngeal neisseriae, but the possible role of other genetic mechanisms cannot be entirely ruled out (9).
Surveillance studies have been conducted in Italy since 1994, and a changed pattern of penicillin susceptibility among circulating meningococci during the year 2002 has been observed. In fact, a greater number of PenI strains were detected for which MICs were higher than those found in previous years. A unique clone, C:2b:P1.5, belonging to the ST8/A4 cluster, accounted for 38% of PenI strains isolated in 2002, marking a change in the situation previously observed in Italy (7).
The development of molecular approaches for the prediction of meningococcal susceptibility to penicillin is particularly useful for rapid screening and for the achievement of good interlaboratory standards.
Studying the sequence alignments of our PenI strains, we found the presence of five different amino acid substitutions, derived from exogenous DNA, at the 3′ end of the penA gene. Interestingly, these substitutions were detected in all the penA genes sequenced from PenI meningococci, including those deposited by other authors in GenBank. The real-time PCR assay that we subsequently developed allows one to look for a single genetic target in the prediction of different penicillin susceptibility phenotypes in N. meningitidis. One of the five substitutions had a nucleotide sequence favorable for the design of probes for the real-time PCR in a LightCycler assay. One of the two probes harbors this mutation at the second nucleotide position and successfully identified meningococci as intermediate in agreement with MICs higher than 0.06 μg/ml. Three different melting profiles were provided within the PenI phenotype due to other mismatches in the penA gene sequence. In all cases, the Tm difference between a susceptible and an intermediate strain ranged from 6 to 9.5°C, resulting in discernible and easily interpretable results.
In conclusion, this real-time PCR protocol makes it easy to detect and characterize the penicillin phenotypes of meningococci in a few hours without DNA sequencing. The reproducible results in the detection of penicillin-intermediate N. meningitidis strains underline the reliability of this assay for discriminating among meningococcal isolates. Thus, it could be an extremely useful tool for those long-term monitoring programs which already exist in several countries.
Acknowledgments
We thank the microbiologists of the hospital laboratories participating in the Italian National Surveillance of Bacterial Meningitis for isolating the strains and sending them to the reference laboratory of the Istituto Superiore di Sanitá (Rome). We also thank Tonino Sofia for technical assistance.
This research was partially supported by projects 9B2/C and2012/RI of the Istituto Superiore di Sanitá.
REFERENCES
- 1.Antignac, A., P. Kriz, G. Tzanakaki, J. M. Alonso, and M. K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47:285-296. [DOI] [PubMed] [Google Scholar]
- 2.Arreaza, L., B. Alcalá, C. Salcedo, L. de La Fuente, and J. A. Vázquez. 2003. Dynamics of the penA gene in serogroup C meningococcal strains. J. Infect. Dis. 187:1010-1014. [DOI] [PubMed] [Google Scholar]
- 3.Bowler, L. D., Q.-Y. Zhang, J.-Y. Riou, and B. G. Spratt. 1994. Inter-species recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in Neisseria meningitidis: natural events and laboratory simulation. J. Bacteriol. 176:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon, J. R., M. Pauzé, and K. H. Young. 1983. Spread of penicillinase-producing and transfer plasmids from the gonococcus to Neisseria meningitidis. Lancet i:779-781. [DOI] [PubMed] [Google Scholar]
- 5.Jones, D. M., and E. M. Sutcliffe. 1990. Meningococci with reduced susceptibility to penicillin. Lancet 335:863-864. [DOI] [PubMed] [Google Scholar]
- 6.Kyaw, M. H., J. C. Bramley, S. Clarke, P. Christie, I. G. Jones, and H. Campbell. 2002. Prevalence of moderate penicillin resistant invasive Neisseria meningitidis infection in Scotland, 1994-9. Epidemiol. Infect. 128:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastrantonio, P., P. Stefanelli, C. Fazio, T. Sofia, A. Neri, G. La Rosa, C. Marianelli, M. Muscillo, M. G. Caporali, and S. Salmaso. 2003. Serotype distribution, antibiotic susceptibility, and genetic relatedness of Neisseria meningitidis strains recently isolated in Italy. Clin. Infect. Dis. 36:422-428. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing: 11th information supplement. NCCLS document M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Orus, P., and M. Vinas. 2001. Mechanisms other than penicillin-binding protein-2 alterations may contribute to moderate pencillin resistance in Neisseria meningitidis. Int. J. Antimicrob. Agents 18:113-119. [DOI] [PubMed] [Google Scholar]
- 10.Peyret, N., P. A. Seneviratne, H. T. Allawi, and J. SantaLucia, Jr. 1999. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A-A, C-C, and T-T mismatches. Biochemistry 38:3468-3477. [DOI] [PubMed] [Google Scholar]
- 11.Richter, S. S., K. A. Gordon, P. R. Rhomberg, M. A. Pfaller, and R. N. Jones. 2001. Neisseria meningitidis with decreased susceptibility to penicillin: report from the SENTRY antimicrobial surveillance program, North America, 1988-99. Diagn. Microbiol. Infect. Dis. 41:83-88. [DOI] [PubMed] [Google Scholar]
- 11a.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spratt, B. G., and K. D. Cromie. 1988. Penicillin-binding proteins of gram-negative bacteria. Rev. Infect. Dis. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 13.Spratt, B. G., Q.-Y. Zhang, D. M. Jones, A. Hutchinson, J. A. Branningan, and C. G. Dowson. 1989. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 86:8988-8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tettelin, H., N. J. Saunders, J. Heideleberg, et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, Q.-Y., D. M. Jones, J. A. Saez-Nieto, E. Perz Trallero, and B. G. Spratt. 1990. Genetic diversity of penicillin-binding protein 2 genes of penicillin-resistant strains of Neisseria meningitidis revealed by fingerprinting of amplified DNA. Antimicrob. Agents Chemother. 34:1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]