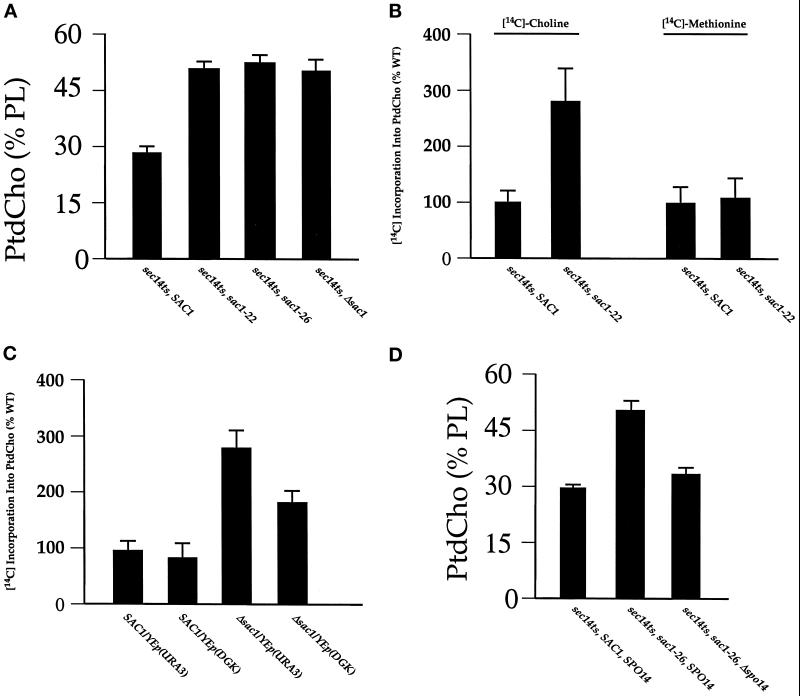
Figure 3.
Phosphatidylcholine synthesis in wild-type and sac1 mutant yeast strains. (A) Strains with the indicated genotypes (at bottom) were grown to midlogarithmic growth phase in medium containing inositol (0.1 mM) and choline (1 mM). Cell cultures were then pulse radiolabled with [32P]orthophosphate (10 μCi/ml) for 30 min at 25°C. Phospholipids were extracted by the method of Atkinson (1984) and resolved by two-dimensional chromatography, and radiolabeled species were quantified by phosphorimaging (see MATERIALS AND METHODS). Biosynthetic rates were deduced from the relative proportion of label recovered in each individual phospholipid species (expressed as percentage of total phospholipid). Rates of total phospholipid synthesis in these strains were estimated by measuring incorporation of 32P into chloroform-soluble counts (11,300–12,600 cpm/OD600 for sac1 and sac1, Δspo14 mutants to 10,000–11,000 cpm/OD600 for wild-type strains). Error bars are indicated. Phospholipid species are presented in the following order: PtdIns (black bars), PtdCho (cross-hatched bars), PtdSer (hatched bars), PtdEtn (stippled bars), and phosphatidic acid (open bars). Strains used were CTY1-1A (sec14-1ts, SAC1, SPO14), CTY165 (sec14-1ts, sac1-22, SPO14), and CTY1261 (sec14-1ts, sac1-22, Δspo14). These data represent an average of three independent experiments, and all strains were analyzed in parallel for each trial. In a typical quantitation, PtdCho accounted for 5.0 × 105 and 2.7 × 105 phosphorimager units in sac1 and SAC1 strains, respectively, when 1 × 106 phosphorimager units of total phospholipid were analyzed from each strain. (B) Activities of the CDP-choline pathway and PtdEtn methylation pathway were evaluated in vivo by pulse radiolabeling of SAC1 and sac1-22 strain pairs (CTY1-1A and CTY165, respectively). The indicated strains were grown to midlogarithmic growth phase in choline-free medium containing inositol. Cell cultures were split, and one aliquot was pulse radiolabeled with [14C]choline chloride (1 μCi/ml) or [14C]methyl-methionine (1 μCi/ml), respectively, for 20 min at 26°C with shaking. A parallel aliquot was labeled with [32P]orthophosphate for 20 min at 26°C. Phospholipids from equal cell equivalents (determined by 32P incorporation into phospholipid in the parallel cultures) were subsequently extracted, resolved, and quantified as described in MATERIALS AND METHODS. These data represent an average of three independent experiments. For quantitative comparison of a representative experiment, wild-type and sac1 strains incorporated 3.8 × 104 and 1.3 × 105 cpm [14C]choline chloride into PtdCho per OD600 cells, respectively. (C) Strains with the indicatedgenotypes were grown to midlogarithmic growth phase in choline-free medium containing inositol. Cell cultures were split, and one aliquot was pulse radiolabeled with [14C]choline chloride (1 μCi/ml) or [32P]orthophosphate (10 μCi/ml) for 20 min at 26°C. Phospholipids were extracted, resolved, and quantified (see MATERIALS AND METHODS). Again, samples were normalized for cell equivalents based on incorporation of [32P]orthophosphate into the parallel cultures as described in B. These data represent an average of three independent experiments. Strains CTY182 (SAC1) and CTY244 (Δsac1) were transformed with the test YEp(DGK) plasmid and the vector-only control YEp(URA3) plasmid, respectively. The YEp(DGK) plasmid was represented by the pCTY85 construct that drives constitutive expression of DGK in yeast (Kearns et al., 1997).