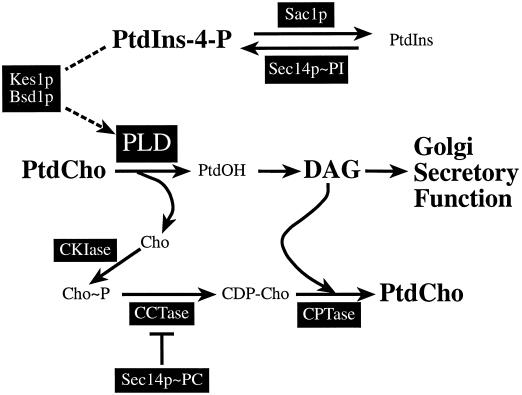
Figure 8.
Bypass Sec14p mechanisms. The relevant lipid metabolic pathways and proteins that determine the activity of these pathways are shown. The key lipids (i.e., PtdIns-4-P, PtdCho, and DAG) are indicated in bold. Key proteins and their corresponding sites of action are indicated in the black boxes. We propose two principal mechanisms for bypass Sec14p. First, we suggest that Kes1p, SacIp, and Bsd1p are involved in a PLD regulatory pathway that, in yeast mutants carrying the corresponding bypass Sec14p alleles, results in PLD activation and subsequent production of DAG. This particular pathway recapitulates the physiological function for the PtdIns-bound form of Sec14p (Sec14p∼PI) that we have proposed effects DAG production via regulation of inositol phospholipid metabolism (Kearns et al., 1997). Loss of SacIp function results in PtdIns-4-P accumulation that, in turn, either activates Bsd1p (identified by dominant bypass Sec14p alleles; Cleves et al., 1991b; Fang et al., 1996) or inactivates Kes1p (identified by recessive bypass Sec14p alleles; Fang et al., 1996). The consequence is activation of PLD, which results in DAG production through the metabolism of the PtdOH generated by PLD-mediated hydrolysis of PtdCho. We propose that the excess DAG generated via this route is consumed by the CDP-choline pathway for PtdCho biosynthesis, thereby resulting in the observed activation of this pathway biosynthetic pathway in sac1 mutants. Second, we suggest that dysfunction of any one of the enzymes of the CDP-choline pathway results in the observed recessive bypass Sec14p phenotypes (Cleves et al., 1991b), because inactivation of the CDP-choline pathway recapitulates the function of PtdCho-bound Sec14p (Sec14p∼PC) (Skinner et al., 1995). Although bypass Sec14p via this route also requires PLD activity, there is nonetheless a measurable PLD-independent component to suppression of Sec14p defects by CDP-choline pathway defects.