Figure 1. Mutation of the LBR locus in EML/EPRO-ic/ic cells inhibits LBR transcriptional activation during neutrophil differentiation.
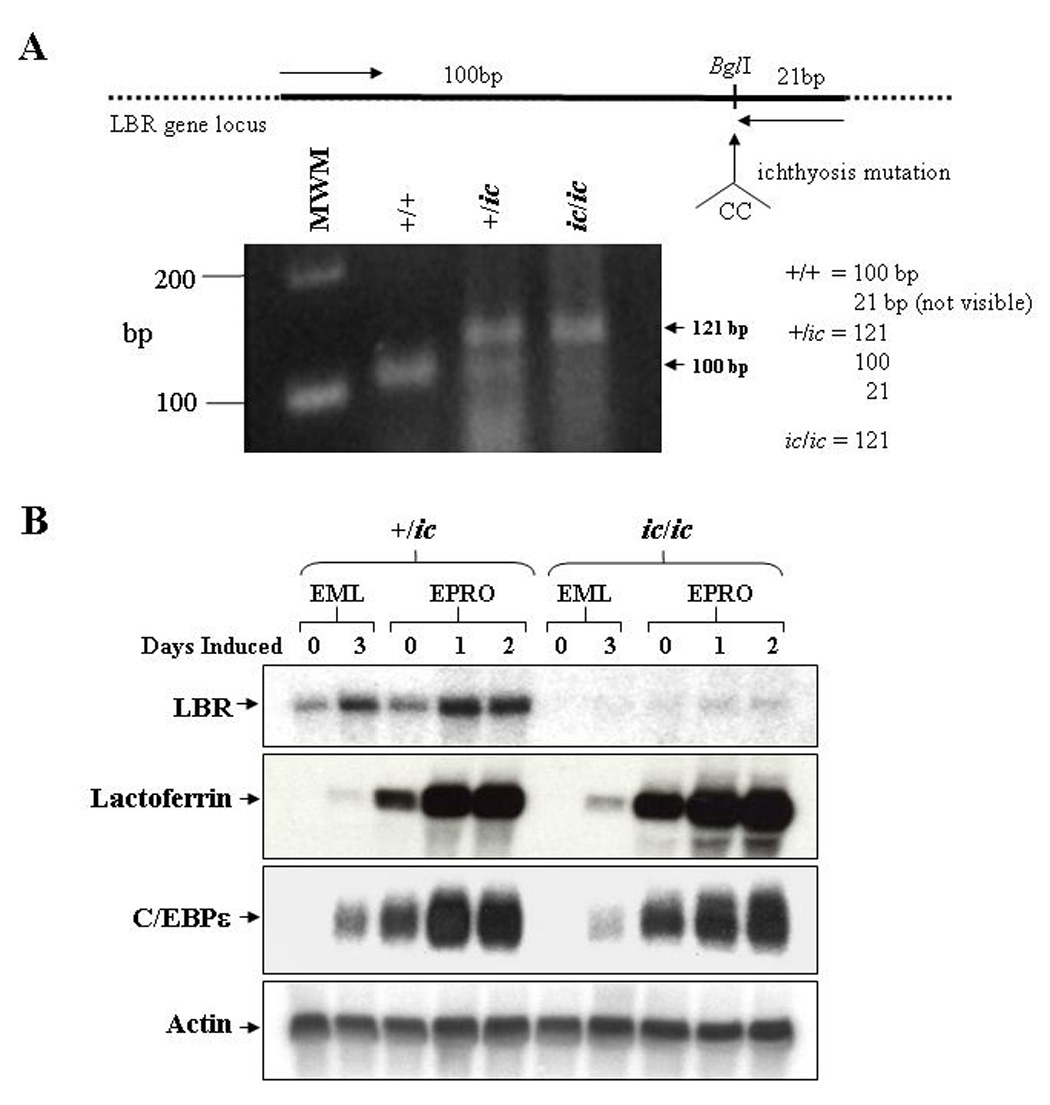
(A) Genomic analysis of the derived EML cell lines identifies the ichthyosis mutation. Genomic DNA from each genotype was isolated and subjected to PCR amplification using primers (indicated by arrows in the upper diagram) specific to the LBR gene but that create a BglI site at the 3’ end of the fragments. This site is disrupted by the ichthyosis mutation (CC insertion). Digestion of the amplified 121bp fragments from wild-type cells with BglI yield 100 and 21bp fragments, cells with the ichthyosis mutation yield only 121bp fragments, and cells heterozygous for ic yield all three fragments. Shown below the diagram are the amplified products from each genotype that were electrophoresed and stained with ethidium bromide. (B) Expression of LBR and neutrophil-specific genes in differentiated ic/ic cells versus +/ic cells were analyzed by Northern blot hybridizations. Total RNA (10 µg) from undifferentiated vs. ATRA-induced EML/EPRO cells of each genotype was electrophoresed, blotted and sequentially probed with labeled cDNA for lactoferrin, C/EBPε and actin as a loading control. The changes in LBR expression shown in each genotype are representative of at least three separate inductions and northern analyses.
