Abstract
Antisense oligodeoxynucleotides (AONs) and short interfering RNAs (siRNAs) effect posttranscriptional gene silencing (PTGS) by hybridizing to an mRNA and then directing its cleavage. To understand the constraints that mRNA structure imposes on AON- vs. siRNA-mediated PTGS, AON- and siRNA-mediated cleavage of defined mRNA structures was monitored in Drosophila embryo whole-cell lysates. We observed that AON-directed cleavage was ≈3-fold faster than cleavage with a siRNA directed to the same target site. Furthermore, and unexpectedly, AON-mediated cleavage was found to be much less fastidious with respect to target sequence accessibility, as measured by the presence of unpaired nucleotides, than a corresponding siRNA. Nonetheless, in vivo, siRNAs silenced their mRNA target at least 2-fold more efficiently than the corresponding AON. These seemingly contradictory results suggested that additional, as yet undefined factors play an important role in regulating PTGS efficiency in vivo. We used a well defined RNA-binding protein, αCP, and its corresponding high-affinity RNA-binding site to explore this hypothesis. We found that prebound αCP effectively blocked AON-mediated cleavage of the RNA-binding site compared with cleavage of the site in the absence of αCP. We conclude that higher-order structures formed by RNA and bound proteins play an important role in determining the efficiency of AON-directed PTGS. We hypothesize that strategies aimed at removing RNA-binding proteins might significantly improve AON-mediated PTGS in vivo.
Keywords: antisense, oligodeoxynucleotide, siRNA
Impairing protein synthesis by disrupting the ability of a cell to use messenger RNA (mRNA) is termed posttranscriptional gene silencing (PTGS). PTGS is a natural phenomenon that can be accessed in the laboratory through the use of single-stranded antisense oligodeoxynucleotides (AONs), ribozymes, DNAzymes, or short interfering RNA (siRNA), all of which may direct degradation, or block translation, of the mRNA to which they are targeted (1, 2). The function of hundreds of genes has been inferred by using PTGS in the laboratory. Beyond this use, many hope PTGS will make a significant contribution to the therapy of a wide variety of diseases associated with the aberrant expression of specific genes (3, 4).
Oligonucleotide-induced PTGS depends on heteroduplex formation with the targeted mRNA. It is intuitive that higher-order mRNA structure might perturb this process, and objective evidence supports this supposition (5, 6). For example, in a study targeting the HIV-1 transactivation response element (TAR), it was found that disruption of the characteristic TAR hairpin resulted in greatly enhanced siRNA activity (7). In studies of a similar nature, it was generally concluded that siRNA efficacy increased commensurately with target sequence accessibility (8–10). Because it is difficult to predict sequence accessibility in vivo (11), numerous chemical modifications have been made to siRNAs and AONs to augment their binding affinity and help overcome the hybridization barriers posed by mRNA structure (12, 13). Nonetheless, this strategy and others have not alleviated the problem of targeting mRNA with oligonucleotides. Part of this difficulty is attributable to the fact that knowledge of the mechanistic details governing oligonucleotides recognition and duplex formation with their mRNA target remains incomplete. In particular the mRNA structural characteristics that impact hybridization are not well defined, and with specific regard to this report, to the best of our knowledge no study has compared the in vitro activity of the RNase H-based mechanisms of AON and siRNA by monitoring the actual mRNA cleavage event.
To formally address the issue of RNA structure and its effects on AON- and siRNA-mediated PTGS pathways, we developed an in vitro assay to monitor the cleavage of a fixed sequence target within a 182-nt RNA molecule. Fixing the sequence of the targeting oligonucleotides was a critical element of our model system because it is known that this is a variable effecting PTGS efficiency (14, 15). With sequences held constant, under the reaction conditions described, higher-order structure of the target mRNA, which was imposed at defined positions by pre-annealing noncleavable 2′-O-methyl oligonucleotides (2′OMe ON), became the sole variable affecting cleavage. Our studies revealed, surprisingly, that in vitro, AON cleave an identical RNA target at a rate approximately three times faster, and with less regard to nucleotide base pairing within the target, than a corresponding siRNA. This advantage was not maintained when attempting to cleave the same target in vivo. Our data suggest that local protein binding may explain this discrepancy, at least in part, highlighting the continuing challenge of identifying, and manipulating, the critical variables impacting PTGS targeting strategies.
Results
Direct Comparison of RNase H- and RISC-Induced mRNA Cleavage Activity.
Before initiating studies on the relationship between mRNA substrate structure and PTGS, a direct comparison of the enzymatic activity of RNase H and RISC was deemed necessary because, to the best of our knowledge, no such comparison is available in the literature. To conduct this investigation, we adapted an in vitro RNA cleavage assay employing a Drosophila embryo whole-cell lysate rich in RISC activity (16). For ease of analysis, and to increase nuclease resistance, we used a 182-nt segment of firefly luciferase with a 5′ radioactive cap (17). To facilitate direct comparison, the AON was identical in sequence to the siRNA guide strand used for the cleavage reactions. Maximal loading of the siRNA into the RISC was assured by preincubating the siRNA in the lysate before the addition of the mRNA target. Although AON activity is not thought to depend on incorporation into a multiprotein complex, AONs were also preincubated in lysate for the same amount of time before the addition of the mRNA target.
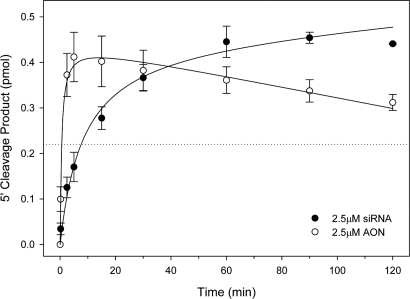
Reactions conducted with either 2.5 μM AON or 2.5 μM siRNA demonstrated considerable activity as evidenced by the efficient generation of the expected ≈104-nt product [Fig. 1 and supporting information (SI) Fig. S1]. In the first 2.5 min of the reaction, the AON and siRNA generated an average of 0.37 and 0.13 pmol of 5′ cleavage product equating to an initial rate difference of ≈2.8-fold (Fig. 1). After 30 min of reaction time, however, both AON and siRNA generated similar amounts of specific 5′ cleavage product. Presumably due to a coincidental repeat of GAAUAC in both the 19-nt target site and upstream in the body of the mRNA, the AON also generated a 51-nt secondary cleavage product that is not observed in siRNA reaction (Fig. S1a). Based on these results, we elected to examine the effects of mRNA structure on cleavage activity in the subsequent experiments by assessing AON- and siRNA-mediated product formation at static time points of 30 s and 8.5 min, respectively (Fig. 1).
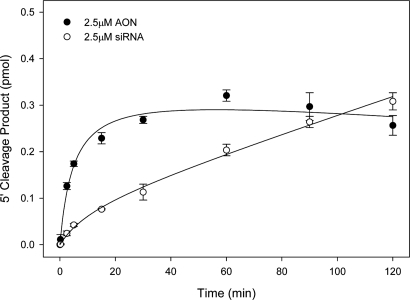
Fig. 1.
Comparison of AON and siRNA activity in Drosophila embryo whole-cell lysate. A 182-nt 5′-cap-labeled segment of firefly luciferase mRNA was incubated in Drosophila embryo whole-cell lysate with 2.5 μM AON or siRNA. Aliquots were removed at 0, 2.5, 5, 10, 15, 30, 60, 90, and 120 min and analyzed by sequencing gels and phosphor-imagery. The mean of 5′ cleavage product formation as a function of time is plotted (at least three independent reactions, ±1 SD). The dotted line at 0.22 pmol product indicates single time points used in later experiments to compare AON and siRNA activity against structure targets.
Effect of Imposing Structure Upstream and Downstream of the Target Site on AON- and siRNA-Mediated mRNA Cleavage in Vitro.
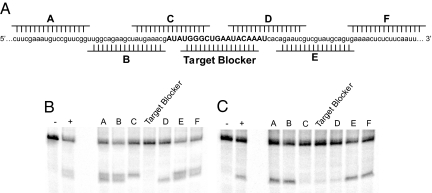
With variables such as oligonucleotide delivery, siRNA incorporation into RISC, GC content, and TM controlled for, we then explored the effect of RNA structure on the rate and efficiency of oligonucleotide-directed mRNA cleavage. We began these studies by examining the relationship between mRNA cleavage and the imposition of known structure at various sites along the mRNA being targeted. This was accomplished by annealing 2′OMe ON at specific sites along the mRNA relative to the target site before in vitro cleavage reactions (Fig. 2A and Table S1, Group I). The 2′OMe ON, used because of their high affinity for mRNA and inability to support RNase H cleavage (18), formed short duplexes either upstream (2′OMe A, B) or downstream (2′OMe E, F) of the targeted sequence within the mRNA. The 2′OMe ON “Target Blocker” was a perfect complement to the actual target sequence and when hybridized created a short, 19-nt double-stranded target. Upstream and downstream regions of the target were made double-stranded with 2′OMe ON C and D, respectively (Fig. 2A). All 2′OMe ON used were annealed to mRNA at a 1.5:1 molar ratio. Under final assay conditions, the AON and siRNA were each used at concentrations >15-fold higher than the 2′OMe ON.
Fig. 2.
Analysis of inducing structure over a large area of the 182-nt luciferase mRNA target. Partial sequence of mRNA target is shown (A) with the relative position of each individual pre-annealed 2′OMe ON A–F. The target sequence of the AON and siRNA guide strand is indicated in all capital letters. Each induced structure of A was used in cleavage assays for 30 s in the presence of 2.5 μM AON (B) or for 8.5 min in the presence of 2.5 μM siRNA (C) and analyzed on 10% sequencing gels. Negative controls (−) have no siRNA or AON, and positive controls (+) are cleavage reactions in the absence of 2′OMe ON.
When flanking 2′OMe ON A, B, E, or F were pre-annealed to the mRNA, cleavage of the mRNA target was actually enhanced by both the AON (measured at 30 s) and siRNA (measured at 8.5 min) (Fig. 2 and Fig. S2). The latter finding is consistent with a previous report that shows enhanced siRNA activity after disruption of local mRNA structure (7). In reactions where half of the target site was complexed with 2′OMe ON C or D, siRNA activity was hindered to a significantly greater extent than AON. Interestingly, however, when the target site was completely blocked by annealing the 2′OMe Target Blocker, the siRNA's activity was reduced by approximately the same amount as reactions carried out with 2′OMe C and D. In contrast, cleavage induced by the AON was reduced to the level of the negative controls (Fig. S2). We also noted that the siRNA generated a consistent cleavage product regardless of where the 2′OMe ON was pre-annealed to the target. In contrast, when cleavage was directed by an AON, blocking either the 5′ (2′OMe C) or 3′ (2′OMe D) end resulted in the RNase H cleavage site shifting in the 5′ or 3′ direction accordingly. These data are consistent with the observations that RISC is a site-specific nuclease (19), whereas RNase H is more promiscuous (20). These observations emphasize that even when processing the identical substrate, these enzymes, which share similar nuclease domains, recognize and process their substrates quite differently.
RNA Cleavage as a Function of Base Pairing Within the Target Sequence.
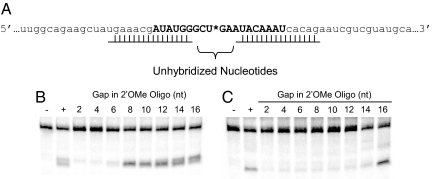
We next sought to more clearly define the target access requirements for cleavage reactions mediated by AONs and siRNAs. To vary target-site accessibility, double-stranded regions involving a lesser or greater number of nucleotides within the target site were created by pre-annealing Group II 2′OMe ONs (Table S1). The target site was encroached on in a symmetrical manner such that an equal number of unpaired bases always remained on either side of the nucleotide where the actual cleavage reaction occurred (Fig. 3A).
Fig. 3.
Determination of the minimum number of accessible bases for AON- and siRNA-guided cleavage in vitro. (A) Partial sequence of the 182-nt firefly luciferase target mRNA. Two 2′OMe ON were annealed at the outer edges of the target site (all capital letters) and positioned incrementally closer to the expected siRNA cleavage site (asterisk). Analysis of the 30-s AON (B) and 8.5-min siRNA (C) reactions was performed on 10% sequencing gels where negative controls (−) have no AON or siRNA and positive controls (+) target the mRNA in the absence of 2′OMe ON.
After 30 s, AON-mediated cleavage of the target RNA was maintained or slightly enhanced compared with that observed in the positive control reaction (AON plus Target RNA alone), when as little as 8 nt of the target site remained unpaired, and therefore “accessible” (Fig. 3B). It was also noted that multiple RNase H-generated cleavage products were generated when 14 and 16 nt of the 19-nt target region was unpaired, whereas fewer products were observed when the target site was more constrained by exposing only 8, 10, or 12 nt of the target (Fig. 3B). When 2′OMe ONs were used to reduce target accessibility to 6 nt or less, AON activity ranged from 56% to 27% of the positive control (Fig. 3B and Fig. S3).
In contrast to these results, the 8.5-min siRNA cleavage reactions generated a consistent single reaction product (Fig. 3C). When compared with the positive control, siRNA-directed cleavage was maintained or even slightly enhanced when 16 nt of the target was left accessible and flanked by the hybridized regions (Fig. 3C). However, when any less than 16 nt was left unpaired, siRNA activity decreased dramatically, ranging from 52% to 29% of the positive control (Fig. 3C and Fig. S3).
Effect of Imposing Structure Within the Target Site on AON- and siRNA-Mediated mRNA Cleavage.
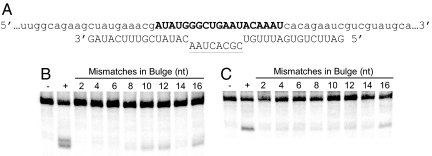
Having examined the influence of unpaired bases within the target site on AON- and siRNA-mediated mRNA cleavage, we next attempted to further our understanding of higher-order target-site parameters by imposing known structure within the target site itself. The Group III 2′OMe ONs (Table S1) were designed such that the 15-nt wings of the 2′OMe ON paired perfectly with the mRNA while the interior sequence was designed to yield base pair mismatches ranging between 2 and 16 nt in length. This strategy was designed to create bulges of defined size, centered around the expected bond of siRNA-induced cleavage, within the otherwise double-stranded target-site region (Fig. 4A).
Fig. 4.
Analysis of cleavage activity when targeting a double-stranded mRNA with a variable internal loop. (A) Partial sequence of the 182-nt firefly luciferase target. One 2′OMe ON (plain capital letters) of differing length was pre-annealed over the center of the target site (bold capital letters). Two 15-nt reverse complementary arms were always annealed to the target to maintain duplex stability. Cleavage activity is reported as a function of the number of mismatches (underlined capital letters) used to vary the size of the internal loop. Analysis of the 30-s AON (B) and 8.5-min siRNA (C) reactions was performed on 10% sequencing gels where negative controls (−) have no AON or siRNA, and positive controls (+) target the mRNA in the absence of 2′OMe ON.
In comparison with the previously examined structures, the entire set of double-stranded targets greatly reduced AON- and siRNA-mediated cleavage activity (Fig. 4). For both AON and siRNA, activity was maximal with the largest bulge and decreased predictably as the size of the bulge progressively diminished (Fig. S4). The lone, but consistent, exception to this rule was observed with an internal loop of 14 nt that exhibited a greater than expected decrease in cleavage product. The reason for this remained unclear.
To determine whether target sites in the “bulge” could be invaded over time by either the RISC or AON, the cleavage reactions with 6-nt bulged targets were extended for up to 2 h. At 2.5, 5, 15, 30, 60, 90, and 120 min, the reactions were sampled and the products analyzed by gel electrophoresis. The primary cleavage product generated by the AON was initially a single band, but after 15 min additional cleavage products were generated (Fig. S5). Additionally, the AON generates the secondary cleavage product seen previously (Fig. 1A) at a similar rate as the primary cleavage product. The siRNA began to generate a single primary cleavage product visible after 15 min (Fig. S5). This continued to accumulate throughout the sampling period so that by 2 h the amount of cleavage product generated by the siRNA was virtually identical to that produced by the AON (Fig. 5).
Fig. 5.
Cleavage activity as a function of time with double-stranded target. Two-hour cleavage reactions in the presence of 2.5 μM AON or siRNA are analyzed on 10% sequencing gels. Primary cleavage product formation as a function of time is reported using the mean of at least three independent reactions (±1 SD).
In comparison with reactions targeting the native mRNA structure (Fig. 1), the total amount and rate of primary cleavage product formation was decreased ≈50% for both AON and siRNA (Fig. 5). The comparative kinetics of the cleavage reactions was not dissimilar to those observed with the native target in that the amount of AON generated product rose rapidly and then plateaued. In contrast, the siRNA-generated product began to accumulate slowly but at a relatively constant rate that was still proceeding when the reaction was terminated. These results suggest that both AON- and siRNA-mediated cleaving complexes have the ability to deal with significant levels of target-site structure over time. Interestingly, it also appears that once access is gained, cleavage proceeds as if the target site were uninvolved by structure.
AON- vs. siRNA-Mediated PTGS Compared in Vivo.
The preceding results might suggest that if the oligonucleotides described above were targeted against the identical target in vivo, the AON would yield cleavage product more quickly and in amounts equivalent to or greater than those generated by the siRNA. This presumption was tested in K562 chronic myelogenous leukemia cells, a cell line that is efficiently transfected with multiple plasmids (21). Contrary to expectations, when firefly luciferase mRNA was expressed and then targeted in K562 cells, a reduction in relative luminescence of 35% and 78% was observed for the AON and siRNA, respectively (Fig. S6). Scrambled sequence AON and siRNA controls showed no effect on the activity of the luciferase reporter construct compared with activity of the vector alone. Therefore, the AON′s rate of cleavage advantage in vitro, in comparison with the siRNA, did not accurately predict which of the two targeting approaches would be most efficient in intact cells. Therefore, some factor, or set of factors, either impairs AON or facilitates siRNA enzymatic activity, or both, in vivo.
Effect of RNA-Binding Proteins on AON-Directed mRNA Cleavage.
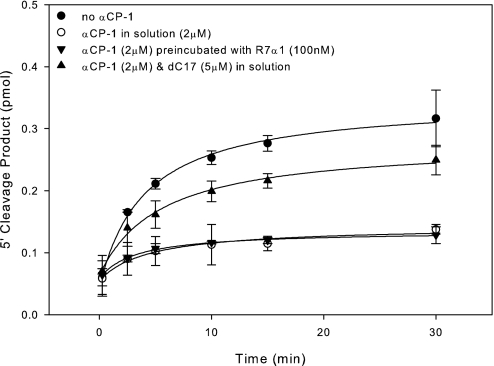
A major structural element so far unaccounted for when targeting an endogenously transcribed target is the effect of RNA-binding proteins. Due to the density at which they bind transcripts (22), RNA-binding proteins may play a major role in limiting the effectiveness of PTGS in vivo. To test this hypothesis directly, we developed an in vitro cleavage assay targeting a 95-nt stem–loop RNA molecule (R7α1) that is bound within the single-stranded loop with high specificity and affinity by the poly(C)-binding protein αCP-1 (23). Because there was no detectable binding of R7α1 by αCP in the Drosophila embryo whole-cell lysate (data not shown), we conclude that there is little, if any, active αCP in our system. Therefore, recombinant human αCP-1 was added at the same concentration as the AON (2 μM) to saturate the R7α1 (data not shown). Unfortunately, none of the siRNA targeting R7α1 were active, even in the absence of αCP. The rate of AON-directed cleavage of the αCP-binding site in the 32-nt single-stranded loop of R7α1 was then measured in the absence of αCP, when the AON and αCP were allowed to compete for binding, and when αCP was preincubated with R7α1 before addition of the AON (Fig. S7). We found that the presence of αCP under all conditions significantly impaired the rate of AON-mediated cleavage (Fig. 6). Addition of a 17-nt poly(C) ODN αCP inhibitor (dC17) to the competitive binding reaction effectively restored AON activity (Fig. 6). These data reveal a major negative impact of an RNA-binding protein on AON sensitivity and indicate that the observed decrease in AON activity is specifically related to the corresponding RNA–protein interaction.
Fig. 6.
α-Complex formation blocks AON activity targeting R7α1 poly(C) regions. Shown is product formation from a minimum of duplicate experiments using AON to cleave R7α1 in the absence of αCP (filled circles), when αCP was in solution with the AON (open circles), when αCP was prebound to R7α1 (inverted filled triangles), or in the presence of αCP and dC17 (filled triangles). Final concentrations were 100 nM R7α1-WT, 2 μM AON-37, 2 μM αCP-1, or 5 μM dC17.
Discussion
Many hope that RNAi will be the major portal through which gene-silencing therapies enter the realm of clinical medicine. Still, significant obstacles remain to be overcome before siRNA-based pharmaceuticals become a reality. Most of these are quite well known because they are virtually identical to the roadblocks that have encumbered the now more classic AON-mediated gene-silencing approaches. Among these are issues related to delivery, intracellular stability, specificity of silencing, knockdown efficiency, and cost of synthesis.
AONs have certain theoretical advantages with respect to at least some of these problems, and the literature suggests that, if carefully chosen, antisense DNA molecules may cleave their targets as efficiently, and in some cases more efficiently, than siRNA (24). We are interested in understanding the structural variables of the target mRNA that dictate the efficiencies of these PTGS approaches. A number of recent studies have suggested that RNA structure can influence the ability of a siRNA to cleave its target (7, 10, 25), but none of these has simultaneously compared the ability of an siRNA and AON to attack the same defined structure in vitro. This is an important distinction because the use of this model system allows one to directly compare how AON and siRNA resolve, or fail to resolve, the same structural issue. Similarly, the impact of other structure modifying factors, such as RNA-binding proteins, on the ability of each oligonucleotide to facilitate RNase H-based cleavage of the same target has not been examined in this manner. In many cases, the results obtained were not expected and were therefore both interesting and informative.
Although RNase H and RISC are members of the same family and have evolved similar domains for cleavage (26), to the best of our knowledge, their ability to cleave a similar substrate in vitro has not been formally compared. Here, we show that RNase H is abundant in the same lysate shown to have most of the known components of the RNAi pathway (16). Therefore, using the lysate allowed us to compare the nucleases' activities at their endogenous relative stoichiometry. Each enzyme demonstrated considerable activity as evidenced by the formation of large amounts of the expected product with an initial rate difference of ≈2.8-fold in favor of the AON (Fig. 1). Because the siRNA was preloaded into RISC, one cannot ascribe the differences in kinetics to the delay that might be caused by the need to incorporate the siRNA into the RISC complex. One must therefore conclude that, at least in vitro, mRNA/AON heteroduplex formation can occur rapidly and, once formed, is cleaved by RNase H much faster than the comparable reaction by RISC. These results are important because Szakiel and colleagues (27) have repeatedly emphasized that the initial kinetics of capture and cleavage are important elements in efficiency of mRNA cleavage by oligonucleotides.
The next finding of surprise to us was rooted in the in vivo experience shared by many that AONs are apparently much more fastidious with respect to mRNA target selection than siRNA molecules. This is reflected by the relative difficulty, and ease, respectively, in finding an oligonucleotide that effectively silences the intended target. However, when systematically inducing structure within an invariant mRNA target with 2′OMe ON, we found that AON activity proved much less dependent on target accessibility than a corresponding siRNA molecule. This might be explained in part by the fact that RNase H substrate recognition positions the enzyme's catalytic domain 7 bp downstream of the double-strand RNA-binding domain (28) and can therefore effectively target an AON/mRNA heteroduplex consisting of only 7–8 bp. In contrast, RISC appears to require, if not more extensive then certainly more ordered, interactions between the RNA target and RISC (29), an observation that our results support. When using a double-stranded substrate, AON activity varied directly with the size of the loop created. In contrast, siRNA activity appeared to be independent, a finding that might be explained by the importance of annealing the 5′ seed region of the siRNA relative to downstream sequence (25, 29, 30). Furthermore, when targeting the 6-nt bulge target over 2 h, AON activity was significantly reduced compared with cleavage obtained with the native target (Fig. 1), but interestingly, the initial rapid burst of product formation was still observed. The siRNA molecule displayed kinetics quite different from those observed when targeting the native transcript (Fig. 5). Here, we observed a constant rate of reaction that we postulate is a direct result of the structure induced, and not product release, which is commonly regarded as the rate-limiting step in Argonaute activity (30). Because in all of our in vitro experiments the products generated by RISC are identical, it suggests to us that their rate of release should be constant as well. Accordingly, we hypothesize that product release is only rate-limiting for freely accessible substrate structure. When the target structure is more complex, which is almost certainly the case under physiologic conditions, structure displacement and target association are likely to play a more important role in regulating the overall rate.
Furthermore, and in contrast to recent work by Ameres et al. (25), in our model system siRNA-mediated cleavage was equally disturbed when encroaching in on the target site from either a 5′ or 3′ direction (Fig. 2). We speculate that the difference between our findings and those of Ameres et al. relate to the model systems used by each group. Specifically, Ameres et al. conducted their cleavage experiments with a minimal system employing affinity-purified RISC, whereas our model system, which used whole-cell lysates, is likely more complete from a biological point of view—i.e., it likely contains important cis-regulatory proteins not assayed for in the Ameres system. This too is an important point because our results directly suggest what Ameres et al. speculate upon: the importance of additional proteins that function to modulate siRNA-directed cleavage in vivo. At the same time, our data do not challenge the finding that purified minimal RISC, composed of Argonaute and the guide siRNA, is able to cleave substrate independent of any other proteins (25); nor do they challenge the finding of several groups that minimal RISC associates with mRNA nonspecifically by diffusion and that it cannot resolve structure on its own (25, 29, 31). They argue instead that with the clear biochemical understanding of how minimal RISC acquires and cleaves its substrate in vitro, models that are more biologically complex, such as those reported here, should now come to the fore in hopes of further enhancing our understanding of these processes.
The final surprise of our results is really a conundrum. If AONs acquire and efficiently cleave their mRNA target faster, and with much less regard for structure, than a corresponding siRNA, why is it that when directed against the identical target in vivo they perform poorly against the same siRNA whose activity should have been impaired by the structure adopted in vivo by the mRNA (Fig. S6)? As noted above, this comparison of in vitro and in vivo activities points to the presence of unidentified modifying activities present in the intact cell. The studies described herein do not suggest the identity of these proteins, but candidates might include components of the replete RNAi machinery including proteins responsible for RISC loading, the ATP-dependent RNA helicase necessary for product release likely to be found in P-bodies, as well as proteins present in translational initiation complexes, and RNA helicases, such as Dbp5 in the nuclear pore complex whose function is thought to strip proteins from the mRNA as it exists the nucleus (32–34). It is equally clear that because delivery of AON was not a factor in these in vivo experiments (21), the AON′s activity must have been impaired by what are likely other cis-regulator proteins, as was illustrated by results presented in Fig. 6. It is clear that RNA–protein interactions can alter the ability of an AON to acquire and cleave an RNA segment that is otherwise unstructured and highly accessible to single-strand probes (23). Because AONs exert their activity in the nucleus (2), the mRNA they encounter is likely to be differentially bound by proteins (22) than the target encountered by siRNA in the cytoplasm (35). One could therefore reasonably hypothesize that strategies aimed at stripping proteins from a candidate mRNA might well increase the activity of AONs in a dramatic way.
Materials and Methods
RNA Target Preparation.
A 181-nt fragment of firefly luciferase mRNA was transcribed (T7 Megashortscript; Ambion) from a DNA template prepared by PCR from pGL3 vector (Promega), using the primer pair 5′-TAATACGACTCACTATAGGAACAATTGCTTTTACAG-3′ (T7 promoter underlined) and 5′-ATTTAGGTGACACTATAGACCGGCATAAAGAATTGAA-3′ (SP6 promoter underlined) (Integrated DNA Technologies). After DNase treatment, the RNA transcript was purified through size-exclusion chromatographic spin columns according to the manufacturer's protocol (Spin-40; Princeton Separations). A 5′ cap label was incorporated onto the mRNA by incubating guanosine 5′-triphosphate (α-32P; Perkin-Elmer) with guanylyl transferase (Ambion) according to the manufacturer's protocol with subsequent size-exclusion chromatographic spin-column purification (Spin-10; Princeton Separations) to yield the final 182-nt mRNA target with sequence of 5′-GGAACAAUUGCUUUUACAGAUGCACAUA U C G A G G UGAACAUCACGUACGCGGAAUACUU C G A A A U G UCCGUUCGGUU G GCAG A A GCUAUGAAACGAUAUGGGCUGAAUACAAAUCACAGAAUCGUCGUAUG C A G UGAAAACUCUCUUCAAUUCUUUAUGCCGGUCUAUAGUGUCACCUAAAU-3′, where primary and secondary cleavage sites are underlined.
The 95-nt R7α1 mRNA was transcribed (T7 MEGAshortscript; Ambion) from a DNA template generated by PCR from the pVA1-R7α1 vector (36) with the primer pair 5′-TAATACGACTCACTATAGGAGAGGATACTACACGT-3′ (T7 promoter underlined) and 5′-GCCAAGCTTCTCTACATGC-3′. Capping and purification was carried out as above, and the final sequence of R7α1 was verified as 5′-GAGAGGATACTACACGTGGAGTGACCTTCTCAACTTT ATATTCCCT TTACC C T T C CCCCAAGGCACTCCATTGCATGTAGAGAAGCTTGGCG-3′ by using the fmol DNA Cycle Sequencing System as directed by the manufacturer (Promega).
siRNA, AON, and 2′-O-methyl Oligonucleotide Preparation.
siRNA sense strand of 5′-AUAUGGGCUGAAUACAAAUdTdT-3′ and antisense strand of 5′-AUUUGUAUUCAGCCCAUAUdTdT-3′ obtained from Integrated DNA Technologies and phosphorylated with T4 polynucleotide kinase (T4PNK; New England Biolabs) and purified with size-exclusion chromatographic spin columns (Spin-10; Princeton Separations), following each manufacturer's protocol. Single strands of siRNA were annealed in lysis buffer (excluding DTT and protease inhibitor) by incubating at 65°C for 5 min and transferred to ice to yield a 25 μM duplex. AON, with the sequence of 5′-ATTTGTATTCAGCCCATAT-3′, obtained from Integrated DNA Technologies with the five terminal phosphate linkages on either end modified to phosphorothioates. 2′OMe ON for inducing structure on or near target site (Table S1, Group I) were obtained from Integrated DNA Technologies, and 2′OMe ON for tiling target site and inducing the bulged double-stranded targets (Table S1, Group II and III) were obtained from the University of Pennsylvania Cancer Center Nucleic Acid Facility (UPenn NAF). The sequence of the active AON targeting the poly(C) regions in R7α1 (AON-37PS) is 5′-AGGGGTAAAGGGAATATAAAG-3′ and was obtained from UPenn NAF. The dC17 inhibitor of αCP was obtained from Integrated DNA Technologies with a sequence of 5′-CCCCCCCCCCCCCCCCC-3′. The five terminal phosphate linkages on either end were modified to phosphorothioates on both AON-37PS and dC17.
siRNA and AON Cleavage Assay.
Drosophila culture, embryo harvest, and whole-cell lysate (WCL) preparation were performed as described in ref. 37. Annealing and in vitro cleavage assays were adapted from those previously described (7, 35, 37). 2′OMe ON were annealed to the target mRNA at a molar ratio of 1.5 to 1 immediately before cleavage assay as described for siRNA annealing above. Before the addition of the RNA-2′OMe ON duplex, AONs or siRNAs were incubated for 15 min at 25°C with WCL in the presence of ATP regeneration mix. Upon full assembly of cleavage reactions, final concentrations were 2.5 μM AON or siRNA, 100 nM mRNA target, 8 μg/μl WCL protein, 20 mM creatine monophosphate (Sigma), 3.5 mM DTT, 0.14 units/μl RNasin (Promega), 0.75 mM ATP (Ambion), 0.17 mM GTP (Ambion), 0.12 μg/μl creatine phosphokinase (Sigma), and 5 μM yeast mRNA (Sigma). Positive control reactions used RNA treated under annealing conditions in the absence of 2′OMe ON, and negative controls were conducted in the absence of siRNA or AON. Static time point cleavage reactions were allowed to proceed for 30 s or 8.5 min for AON and siRNA, respectively, at 25°C and were stopped upon the addition of 20 μg of proteinase K (FisherBiotech) and 0.5 μmol of EDTA (BioWhittaker). Full time course reactions were assembled as larger volumes and conducted as described above. Individual 5-μl aliquots were removed just after initiation of the reaction and 2.5, 5, 15, 30, 60, 90, and 120 min later and added to tubes containing of 20 μg of proteinase K (FisherBiotech) and 0.5 μmol of EDTA (BioWhittaker). After denaturing in formamide at 70°C, reactions were analyzed directly on 10% sequencing gels. Autoradiography was performed by exposing gels on phosphor screens that were scanned by using a STORM 860 phosphorimager (Molecular Dynamics). Product bands were quantified by using ImageJ (38).
Assays targeting R7α1 were conducted in the same fashion as above, with the final concentrations of additional reagents 100 nM cap-labeled R7α1 mRNA, 2 μM AON, 2 units/μl SUPERaseIn (Ambion), 2 μM recombinant αCP-1, and 5 μM dC17.
Cell Culture and Nucleofection.
K562 cells were cultivated in RPMI medium (GIBCO Invitrogen), supplemented with 10% FBS and 0.5% penicillin/streptomycin and maintained at 37°C in a humidified chamber (95% humidity) and 5% CO2. Culture media were changed according to the growth conditions of the cells, usually every second day.
A firefly luciferase vector, pGL3-Promoter [Promega (catalog no. E1761)], was nucleofected into K562 cells either by itself or along with AON or siRNA directed against luciferase gene by using an Amaxa nucleofector system (Kit V/Program Q-29). Controls such as untreated cells, mock nucleofected cells (absence of AON/siRNA), and a scrambled control (AON and siRNA) were included in the assay. Renilla luciferase [Promega (catalog no. E2261)] was used as an internal control to monitor nucleofection efficiency. Briefly, 1 × 106 cells were washed with PBS, pelletted, and resuspended in 100 μl of nucleofection solution along with luciferase vectors and 0.8 nmol of scrambled control or AON/siRNA against luciferase gene. Cells were harvested 24 h after nucleofection and lysed in passive lysis buffer [Promega Dual Luciferase kit (catalog no. E1960)].
Luciferase Reporter Assay.
Dual luciferase values were measured in a Monolight 2010 (Analytical Luminescence Laboratory). The value of firefly luciferase was normalized to Renilla luciferase control for all assay conditions. The knockdown of firefly luciferase gene was calculated by comparing the vector alone to the presence of AON and siRNA and reported as relative luminescence.
Recombinant αCP-1 Preparation.
Recombinant human αCP-1 protein was expressed in E. coli by transformation BL21-CodonPlus(DE3)-RIL competent cells (Stratagene) with pET28a expression vector carrying the αCP-1 coding region. This vector had an N-terminal hexa-histidine sequence, and fusion αCP-1 protein comprised an additional 34 vector-derived amino acids (39). Expressed 6× His-tagged protein was purified under native conditions by metal chelate affinity chromatography (Ni-NTA agarose column; Qiagen). Purified protein was dialyzed against PBS or Tris·HCl buffer, concentrated by centrifugation in a centricon device, and filtered through a 0.22-μm syringe filter. The concentration of recombinant αCP-1 was calculated by using the Bio-Rad Bradford Protein Assay.
Supplementary Material
Acknowledgments.
This work was supported in part by National Institutes of Health Grants P01-CA72765 and R01-CA101859, the Leukemia and Lymphoma Society of America, and a grant from the Pennsylvania Department of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805781105/DCSupplemental.
References
- 1.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 3.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat Rev Drug Discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gewirtz AM. On future's doorstep: RNA interference and the pharmacopeia of tomorrow. J Clin Invest. 2007;117:3612–3614. doi: 10.1172/JCI34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima WF, Monia BP, Ecker DJ, Freier SM. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- 6.Mir KU, Southern EM. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- 7.Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human RISC. Nat Struct Mol Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 8.Yoshinari K, Miyagishi M, Taira K. Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Res. 2004;32:691–699. doi: 10.1093/nar/gkh221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubert S, Grunweller A, Erdmann VA, Kurreck J. Local RNA target structure influences siRNA efficacy: Systematic analysis of intentionally designed binding regions. J Mol Biol. 2005;348:883–893. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Overhoff M, et al. Local RNA target structure influences siRNA efficacy: A systematic global analysis. J Mol Biol. 2005;348:871–881. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Stein CA. Hybridization prediction gets to first base. Nat Biotechnol. 1999;17:751–752. doi: 10.1038/11680. [DOI] [PubMed] [Google Scholar]
- 12.Braasch DA, et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 13.Elmen J, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds A, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 16.Tuschl T, et al. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir SM, et al. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monia BP, et al. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 19.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Lima W, et al. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol Pharmacol. 2007;71:83–91. doi: 10.1124/mol.106.025015. [DOI] [PubMed] [Google Scholar]
- 21.Kalota A, et al. 2′-deoxy-2′-fluoro-β-D-arabinonucleic acid (2′F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res. 2006;34:451–461. doi: 10.1093/nar/gkj455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 23.Thisted T, Lyakhov DL, Liebhaber SA. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and αCP-2KL, suggest distinct modes of RNA recognition. J Biol Chem. 2001;276:17484–17496. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 24.Vickers TA, et al. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 25.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 26.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 27.Patzel V, Sczakiel G. In vitro selection supports the view of a kinetic control of antisense RNA-mediated inhibition of gene expression in mammalian cells. Nucleic Acids Res. 2000;28:2462–2466. doi: 10.1093/nar/28.13.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lima WF, et al. Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3′-DNA/5′-RNA terminus of the heteroduplex substrate. J Biol Chem. 2003;278:49860–49867. doi: 10.1074/jbc.M306543200. [DOI] [PubMed] [Google Scholar]
- 29.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 31.Ma JB, et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Weirich CS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 34.Gross T, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 35.Martinez J, et al. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 36.Makeyev AV, Eastmond DL, Liebhaber SA. Targeting a KH-domain protein with RNA decoys. RNA. 2002;8:1160–1173. doi: 10.1017/s135583820202808x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 38.Rasband WS. Bethesda, MD: US National Institutes of Health; 1997–2007. http://rsb.info.nih.gov/ij. [Google Scholar]
- 39.Chkheidze AN, et al. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C)-binding protein αCP. Mol Cell Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.