Abstract
There is a growing interest in the mechanisms that control the apoptosis cascade during development and adult life. To investigate the regulatory events that trigger apoptosis in whole tissues, we have devised a genetically encoded caspase sensor that can be detected in live and fixed tissue by standard confocal microscopy. The sensor comprises two fluorophores, mRFP, monomeric red fluorescent protein (mRFP) and enhanced green fluorescent protein (eGFP), that are linked by an efficient and specific caspase-sensitive site. Upon caspase activation, the sensor is cleaved and eGFP translocates to the nucleus, leaving mRFP at membranes. This is detected before other markers of apoptosis, including anti–cleaved caspase 3 immunoreactivity. Moreover, the sensor does not perturb normal developmental apoptosis and is specific, as cleavage does not occur in Drosophila embryos that are unable to activate the apoptotic cascade. Importantly, dying cells can be recognized in live embryos, thus opening the way for in vivo imaging. As expected from the high conservation of caspases, it is also cleaved in dying cells of chick embryos. It is therefore likely to be generally useful to track the spatiotemporal pattern of caspase activity in a variety of species.
Keywords: apoptosis, Drosophila, embryos, Apoliner
“Rien n'est mort que ce qui n'existe pas encore.” Guillaume Apollinaire, Alcools (1913)
During embryogenesis, apoptosis contributes to morphogenesis by sculpting tissues. It also maintains tissue homeostasis by removing mis-specified or superfluous cells. Relatively little is known about the mechanisms that allow such cells to be recognized and tagged for elimination (see ref. 1 for review). As a first step toward elucidating these mechanisms, we sought to devise a reporter assay that would highlight, at the earliest juncture and in live animals, cells earmarked for apoptosis. The molecular drivers of cell death are cysteine proteases of the caspase family. Caspases are divided into two classes: (a) the apical caspases, which trigger the apoptotic cascade, and (b) the effector caspases, which cleave cellular proteins and are thus responsible for the apoptotic phenotype (see ref. 2 for review). Because spurious caspase activity is potentially deleterious, caspases are under tight inhibitory control. For example, in the absence of the caspase inhibitor known as the Drosophila inhibitor of apoptosis protein 1 (DIAP1), which is encoded by the thread gene in Drosophila, every cell of the embryo undergoes apoptosis (3). Thus, the proapoptotic genes located at the H99 locus trigger apoptosis by causing the degradation of DIAP1 (2). DIAP1 is also cleaved by downstream effector caspases, which also leads to its degradation (4).
Results and Discussion
Rationale and Design.
We capitalized on DIAP1 cleavage to design a reporter of caspase activity that is effective in live and fixed tissue and could therefore be used to study the spatial and temporal control of these important enzymes during development and tissue homeostasis. The sensor, which we call Apoliner, involves two fused fluorescent proteins that become separated by caspase activity. Following cleavage, the two moieties become differentially localised because of distinct subcellular localisation signals. Specifically, the sensor comprises, from the amino- to the carboxy-terminus, a transmembrane domain (from mouse CD8), mRFP, a caspase sensitive site from DIAP1, a nuclear targeting signal, and eGFP (Fig. 1A). We reasoned that in live cells, the two fluorescent proteins would reside at the membrane but that in the presence of active caspases, the NLS-eGFP moiety would be released and allowed to accumulate in the nucleus. As a caspase sensitive site, we used a fragment of DIAP1 that is efficiently cleaved by the effector caspases Drice and Dcp1 (4). Importantly, this fragment is cleaved more rapidly than the minimal consensus site (DQVD) because it includes a BIR1 domain, which enhances recognition by caspases (P. M., unpublished data). To prevent degradation of NLS-eGFP by the N-end rule, the two asparagines located downstream of the exact cleavage site (DQVDNN) were replaced (see Materials and Methods). For control experiments, we made a construct that included the same region of DIAP1 except that it contained a single point mutation (DQVANN) that abrogates cleavage (4). This construct will be referred to as “Apomut.”
Fig. 1.
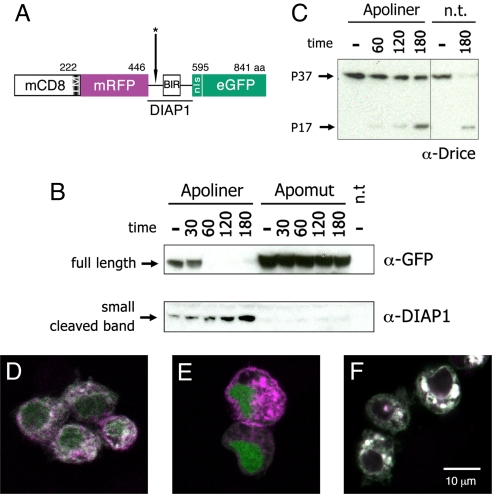
Structure and cleavage of Apoliner. (A) Schematic representation of Apoliner. Numbers indicate position of residues. The cleavage site is marked with an asterisk (see Materials and Methods). (B, C ) Western blots showing Drice, Apoliner, and Apomut at various times following induction of reaper expression in S2 cells. Numbers indicate time in minutes; and minus signs indicate uninduced cells. Apoptosis was induced 48 h after transfection of a vector expressing either Apoliner or Apomut. n.t., no transfection. (B Upper) Western blot stained with antiGFP to detect uncleaved (full length) Apoliner extracted from cells transfected with Apoliner or Apomut. This band has essentially disappeared at 60 min, indicating Apoliner cleavage. (B Lower) As full length Apoliner disappears, a cleavage product, detected with anti-DIAP1 increases. This fragment should also be detectable by anti-GFP, but this is unfortunately masked by a protein of similar size recognized nonspecifically by the anti-GFP antibody (data not shown). Note the absence of any cleavage product in extracts from cells expressing Apomut, a strong indication that Apoliner cleavage depends on the integrity of the caspase cleavage site. Note also that Drice cleavage (appearance of the P17 Drice fragment in C) only begins to be faintly detectable 60 min after induction, a time when full length Apoliner is completely cleaved (as shown in B Upper). (D–F) S2 cells transfected with Apoliner (D, E ) or Apomut (F ), and induced to die by a 10-min ultraviolet exposure. Before induction, RFP (magenta) and GFP (green) colocalize at membranes (giving a white signal in the merge) in a majority of cells (D). After induction, GFP relocalizes to the nucleus of most cells (E; see Fig. S1 for details). Even 180 min after apoptosis induction, cleavage of Apomut could not be detected as indicated by the colocalization of GFP and RFP (F ).
Apoliner cleavage was first assessed on a Western blot. A plasmid encoding Apoliner was transfected in S2 cells carrying MT-reaper (4) so that reaper expression, and hence apoptosis, could be induced at specific times by addition of copper sulfate to the medium. After induction, Apoliner is rapidly cleaved, as reflected by the disappearance of the full-length form in Western blots (Fig. 1B Upper, lanes 2–5). Concomitantly, the GFP-containing fragment can be detected as a band accumulating ≈40 kDa (Fig. 1B Lower, lanes 2–5). Note that this cleavage is already complete at a time when only a faint band of cleaved Drice can be detected (compare Fig. 1B and C, time 60 min). As expected, at this time, nuclear GFP signal can detected by confocal microscopy (compare Fig. 1D and E, and see supporting information [SI] Fig. S1). As a control for the Western blot experiment, we used uninduced MT-reaper cells: only limited cleavage is detected (Fig. 1B, lane 1). Basal cleavage could reflect normal background apoptosis in cultured cells or leaky expression from the uninduced MT-reaper construct. Apoliner cleavage is most likely due to caspase activity since Apomut remains uncleaved even after induction of apoptosis (Fig. 1B, Apomut lanes). Moreover, no GFP is detected in the nucleus of cells expressing Apomut, whether apoptosis is induced or not (Fig. 1F). We conclude that Apoliner is cleaved after activation of apoptotic caspases, suggesting that it is a good caspase sensor.
Apoliner Reports Caspase Activity in Vivo.
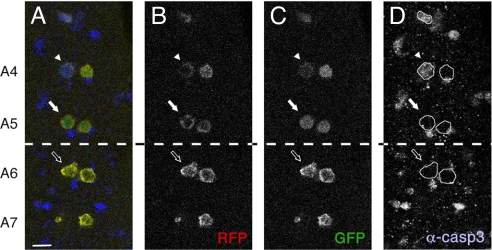
To further assess the effectiveness of Apoliner in vivo, we examined dMP2 neurons, which undergo segment-specific programmed apoptosis in late embryogenesis (5). At embryonic stage 16, two dMP2 neurons are detectable in every segment of the embryonic ventral nerve cord as revealed by expression of the dMP2-Gal4 driver. At embryonic stage 17, dMP2 neurons in the anterior segments (A3 to A5) undergo apoptosis, whereas their counterparts in more posterior segments persist. We used dMP2-Gal4 to express Apoliner and compared the time of cleavage to that of cleaved caspase-3 immunoreactivity in dMP2 neurons. Fig. 2 shows that Apoliner is cleaved in anterior dMP2 neurons (A4 and A5, Fig. 2 A–C, white arrows) but not in the posterior neurons, as expected (A6 and A7, Fig. 2 A–C, outlined arrows). Although dMP2 neurons in anterior segments always die, they activate apoptosis metasynchronously (5). Because of the slight temporal variation, cells at different stages of programmed cell death are seen in a given fixed preparation. This explains the variation in the intensity of anti-activated caspase 3 in A4 and A5 (compare for example the two cells in A4). We find that Apoliner is cleaved in all of the dMP2 neurons that are stained with anti-activated caspase 3. However, the Apoliner signal is always relatively weaker in cells that have strong activated caspase-3 signal (e.g., arrowhead). We suggest that at this late stage of apoptosis, the caspase signal is already waning because of extensive proteolysis. Importantly, Apoliner cleavage is often detected in neurons that have no detectable activated caspase 3 but are known to be fated to die (Fig. 2, white arrow). This suggests that Apoliner identifies apoptotic cells selectively, and before they are recognizable with anti-cleaved caspase 3 antibodies.
Fig. 2.
Apoliner cleavage as seen in the embryonic dMP2 neurons. Ventral nerve cord of a stage 17 Drosophila embryo expressing Apoliner under the control of dMP2-Gal4 (projection of two consecutive confocal slices). RFP (B), GFP (C ) and anti–cleaved caspase 3 (D) are shown independently and in an overlay (A). Apoliner cleavage is restricted to the pairs of neurons in the segments anterior to A6, as expected from the previously reported pattern of apoptosis in these cells (5). Both dying (white arrow and arrowhead) and surviving cells (empty arrow) can be recognized. The caspase-positive cells that are not dMP2 neurons (i.e., not expressing Apoliner) should of course be ignored. (Scale bar, 10 μm.)
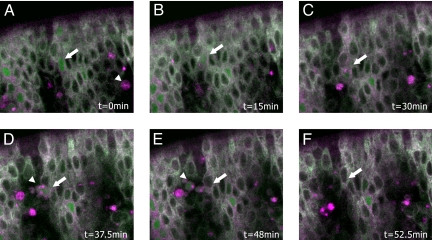
To determine whether Apoliner allows the detection of caspase activity in live embryos, we performed time-lapse imaging of the epidermis, which is optically accessible. In this tissue, there is no precise pattern of cell death but there is a general tendency for apoptotic cells to appear at or around segment boundaries from stage 11 onward (6). Expression of Apoliner in the embryonic epidermis was driven with the 69B-Gal4 driver (7) and the ventral side was imaged by time lapse confocal microscopy from stage 12 onward (i.e., after the onset of developmental apoptosis). Selected frames from a resultant movie (see Movie S1) are shown in Fig. 3. An Apoliner positive cell can be recognized in the first frame (arrow in Fig. 3A) and tracked for an extended period, up to engulfment by a macrophage. Interestingly, macrophages (arrowhead in Fig. 3A, D, and E) become bright red, probably as a result of accumulated membranous RFP from engulfed epidermal cells. The observations above show that Apoliner can be used to track dying cells in live embryos, thus making it a valuable tool to explore the control of apoptosis.
Fig. 3.
Apoliner reports on caspase activity in vivo. Frames from a time-lapse movie tracking an Apoliner-positive cell in a live embryo (see Movie S1). Times are indicated in minutes. Caspase activity is detected by the presence of GFP in the nucleus and the red membrane. One Apoliner-positive cell is marked with an arrow in A–E. In A, this cell appears morphologically normal, although Apoliner is already cleaved. Later, it shrinks (C and D), before being taken up by hemocytes (D and E), and removed from the epithelium. In F, the arrow points to the original position of the removed cell. The arrowhead in D and E points to the hemocyte that engulfed this particular dying epidermal cell.
Apoliner Is Not Cleaved in the Absence of Apoptosis.
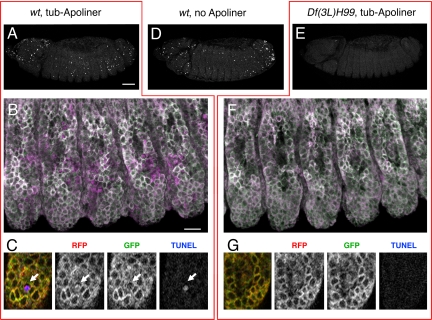
In all of our experiments, we found that cells identified as apoptotic by standard methods (cleaved caspase 3 immunoreactivity, TUNEL staining, the presence of a picnotic nucleus, or engulfment) invariably contain cleaved Apoliner. To ask if Apoliner is spuriously activated in the absence of activated caspases, we expressed Apoliner in H99 mutant embryos, which lack the three major proapoptotic genes (8) and therefore have drastically reduced caspase activity. Such embryos, which are easily recognized by the near complete absence of TUNEL signal (Fig. 4E), show no sign of Apoliner cleavage in the epidermis (see Fig. 4F and compare with control wild-type embryos in Fig. 4B). Similarly, Apoliner cleavage is greatly reduced when p35, an inhibitor of effector caspases (9) is overexpressed (data not shown). These data, along with the finding that Apomut is not cleaved in apoptotic cells (Fig. 1B and F), demonstrate that Apoliner is specifically cleaved upon caspase activation.
Fig. 4.
Comparing Apoliner and TUNEL in normal and apoptosis-deficient Drosophila embryos. (A–C) TUNEL and Apoliner in a control (+/+ or H99/+) embryo. (A) TUNEL signal as seen in a lateral view (projection of 16 consecutive confocal slices). (B) Apoliner in the embryo shown in A, with RFP displayed in magenta and GFP in green, resulting in a white signal upon colocalization. Again, 16 confocal slices are shown, but this time they are projected using the 3D function of Volocity (Improvision). (C) High-magnification view of one confocal slice from B, showing RFP, GFP, and TUNEL, and the overlay on the left-hand side. The arrow points to a shrunken Apoliner-positive cell, which is also positive for TUNEL, an indication of advanced apoptosis. At this late stage, the GFP signal is no longer detectable, presumably because of the high proteolytic activity in this cell (see Fig. S1). (D) TUNEL staining in a wild-type (or H99/+) embryo that does not express Apoliner. Note that the number of TUNEL-positive nuclei is approximately the same as in A, indicating that Apoliner does not interfere significantly with developmental apoptosis. (E–G) TUNEL and Apoliner in a H99-deficient embryo. (E) H99-deficient embryo stained for TUNEL; very little staining can be detected. (F) Apoliner in the embryos shown in E, with RFP in magenta and GFP in green (again, 16 confocal slices projected using the 3D function in Volocity–Improvision). (G) High-magnification view of one confocal slice from F (H99-deficient embryo). In the trunk epidermis, no TUNEL signal or Apoliner cleavage can be detected, whereas an occasional signal is observed in the head region (data not shown; also see ref. 8). [Scale bar for A, D, and E (shown in A), 50 μm; scale bar for B and F (shown in B), 10 μm.]
Apoliner Has No Significant Effect on Apoptosis.
Any biological sensor should have a minimal impact on the process it measures. Three lines of evidence suggest that the expression of Apoliner does not significantly affect caspase activity. First, strong ubiquitous expression of Apoliner (with tub-Gal4) does not affect the number and pattern of TUNEL positive cells in embryos, showing that developmental apoptosis is unaffected (compare Fig. 4A and D). Second, overexpressed Apoliner does not interfere with experimentally induced apoptosis in the eye (see Fig. S2). Third, the timing of apoptosis activation in the dMP2 neurons is unchanged when Apoliner is expressed. As expected then, expression of Apoliner with a variety of Gal4 drivers causes no noticeable effect on organismal viability or health. There is, however, one exception: low-level expression of Apoliner under the direct control of the tubulin alpha1 promoter (10) causes male sterility and morphological defects in the testis (data not shown). Because no deleterious effect is seen in other tissues, the effect seen in the male germ line may result from the disruption of an atypical requirement for caspase activity during spermatogenesis. It is worth mentioning that a BIR1-domain containing protein (dBRUCE) has been shown to have a testis-specific function (11). Note also that Apomut causes the same morphological defects in the testis as Apoliner. It is therefore conceivable that the BIR1 domain, which is present in both constructs, could interfere with dBRUCE function.
Apoliner Is a Reporter of Apoptosis in the Chick.
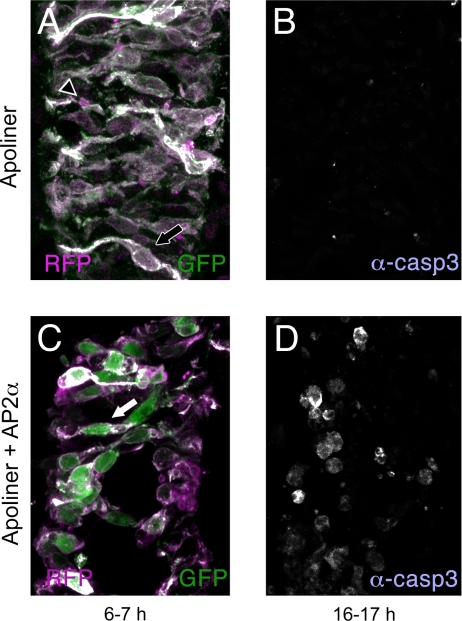
Because caspases are highly conserved, we surmised that Apoliner could be used to reveal caspase activity in other model organisms besides Drosophila. This was tested in chick embryos. A DNA fragment encoding Apoliner was put under the control of the CMV promoter and the resultant plasmid was electroporated into the spinal cord of chick embryos, either alone (Fig. 5A and B) or with an AP2α-expressing construct, which is known to cause apoptosis (M. Cheung and J.B. unpublished observation; Fig. 5C and D). In the absence of experimentally induced apoptosis, 6–7 h after electroporation of Apoliner, the membrane of most transfected cells (e.g., arrow in Fig. 5A) is both magenta (RFP) and green (GFP), appearing white in the merge. Only a few cell corpses are seen (e.g., arrowhead in Fig. 5A). By contrast, 6–7 h after coelectroporation of Apoliner and the apoptosis-inducing plasmid, RFP and GFP no longer colocalize, showing that Apoliner was cleaved (Fig. 5C, arrow). At this early time, relatively few anti–cleaved caspase 3 positive cells are detected (data not shown). Only 10 h after electroporation does widespread anti–cleaved caspase 3 immunoreactivity appear (Fig. 5D), thus confirming the activation of apoptosis detected earlier with Apoliner. We conclude that Apoliner is rapidly cleaved by caspases in chick embryos, and that Apoliner could therefore be used to track caspase activation in this and probably other vertebrates.
Fig. 5.
Apoliner cleavage in the chick spinal cord correlates with increased apoptosis. Each presents a single confocal slice of a chick spinal cord electroporated with either Apoliner alone (A, B), or Apoliner and AP2α (C, D). The spinal cords were fixed either 6–7 h (A, C) or 16–17 h (B, D) after electroporation. Samples were stained with anti–cleaved caspase 3. When only Apoliner is electroporated, few cells are immunoreactive, as shown in B, 16–17 h after electroporation. As expected, both the red and green fluorophores are at the membrane of Apoliner-expressing cells (shown in A, arrow, 6–7 h after electroporation). Relatively few red-only corpses can be seen (outlined arrowhead). These probably reflect the low-level apoptosis that occurs in these samples. By contrast, extensive apoptosis is seen after AP2α expression. This is clearly seen by anti–cleaved caspase 3 staining 16–17 h after electroporation (D) but not yet detectable with the same method 6–7 h postelectroporation (data not shown). In similarly treated spinal cords, as early as 6 h after electroporation, GFP and RFP are seen in distinct subcellular compartment more frequently than in controls (C, arrow). As can be seen, the GFP fills a large fraction of the cell body, consistent with nuclear localization, because the nucleus occupies most of the cell body in these neurons at this stage (see ref. 17 for example).
Conclusion
In summary, Apoliner provides an early marker for caspase activity. The results of our experiments in Drosophila show that Apoliner is specific and relatively innocuous. Because it is genetically encoded, it can be expressed in a subpopulation of cells. By combining this feature with cleavage detection on Western blots, one can envisage tracking cell death in specific tissues in a quantitative way. Furthermore, Apoliner is suitable for live imaging, thus allowing dying cells to be tracked over extended periods of time, potentially from activation of apoptosis to removal by macrophages. Importantly, Apoliner does not require sophisticated imaging equipment as it relies on two commonly used fluorescent proteins. In addition, Apoliner is not merely based on a minimal cleavage site. It contains a recognition sequence that boots efficiency and specificity (see Rationale and Design). These unique features make Apoliner a good alternative to previously described FRET-based sensors (see ref. 12 for example). Our chick experiments suggest that Apoliner will find applications in a variety of model organisms. Although Apoliner primarily marks apoptotic cells, it is important to point out that it is a caspase sensor not an apoptosis marker. It is therefore expected to also detect caspase activity that is not accompanied by apoptosis. Indeed, consistent with earlier reports of nonapoptotic caspase activity (reviewed in ref. 13), we have observed patterned low level Apoliner cleavage in imaginal disk cells that are known not to undergo apoptosis (unpublished data). Apoliner is therefore a general marker of caspase activity that will help exploring how these important enzymes are regulated during development.
Materials and Methods
Generation of the Apoliner and Apomut Constructs.
DNA encoding the N-terminal region (transmembrane domain) of mouse CD8 and eGFP (10) was used as a starting point. The coding region of monomeric RFP (14) was amplified by polymerase chain reaction with primers containing BglII sites and inserted at the unique BamHI site lying in between mCD8 and eGFP. In the 3′ primer, we also incorporated a AgeI, a BamHI site and a DNA fragment encoding the NLS of the large T antigen (CCCAAGAAGAAGCGCAAGGTG). The N-terminal part of DIAP1 (amino acids 1–146) was amplified by polymerase chain reaction and inserted between the AgeI and BamHI sites. The actual cleavage site of DIAP1 (DQVDNN) was modified to DQVDGV to prevent degradation of the cleaved product by the N-end rule pathway (4). This was used to generate Apoliner. In a different construct, we used a mutated fragment of DIAP1 that cannot be cleaved (DQVANN). This was used as a control for the specificity of Apoliner and is called Apomut (Fig. 1 and data not shown). After insertion of the DIAP1 fragment, the DNA encoding Apoliner or Apomut was excised from Bluescript using the XhoI (blunted) and XbaI sites. The resulting fragment was ligated in the NotI (blunted) and XbaI sites of a CaSpeR4 previously modified to carry the tubulin1α promoter and the SV40 polyadenylation site (10). The resulting Tub-Apoliner (and Tub-Apomut) was used for expression in S2 cells and also to make transgenic flies. Apoliner was also introduced in the XhoI and XbaI sites of pUAST to generate UAS-Apoliner. Insertions on the first, second and third chromosomes were recovered but UAS-Apoliner-5, inserted on the second chromosome, was mostly used in this study. Finally, Apoliner was cloned in the pSC2+ vector to put it under the control of CMV promoter for expression in chick embryos (Fig. 5). This is called CMV-Apoliner.
Cell Transfection and Western Blotting.
S2 cells and S2-MT-reaper cells, i.e., stable S2 cells expressing reaper under the control of the metallothionine promoter (15), were grown in Gibco Schneider's medium supplemented with 1% l-glutamine, 1% penicillin/streptavidin, and 10% fetal calf serum. Cells were transfected with 1 μg of Tub-Apoliner or Tub-Apomut using Effectene (Qiagen) according to the manufacturer's instructions. Apoptosis was induced by two different means: either S2 cells were exposed to UV radiation for 10 min, or the expression of reaper was induced 48h after transfection by addition of 0.7 mM CuSO4 to S2-MT-reaper cells. At various subsequent times, the cells were harvested in lysis buffer (50 mM Tris, 2% triton X100, 100 mM NaCl, pH 7.4 plus protease inhibitors (Complete, Roche). Proteins were separated by 4–12% SDS/PAGE (NuPage Novex Bis-Tris gels, Invitrogen). Western blots were probed for drICE, DIAP1, GFP and Actin. The following antibodies were used: anti-GFP (1/2000, rabbit, Abcam, 6556–25), anti-DIAP1 (16) (1/4000, guinea pig) and anti-Drice (16) (1/4,000, guinea pig) and anti-Drosophila actin (1/1000, mouse, JL20, DSHB). Secondary antibodies used were HRP-conjugated anti-rabbit and anti-mouse (1/2,000, Bio-Rad) and anti–guinea pig (Jackson ImmunoResearch, 1/4,000).
Expression in Flies, Immunofluorescence, and Image Acquisition.
Flies carrying UAS-Apoliner-5 were crossed to flies carrying the following drivers: dMP2-Gal4 (5) (Fig. 2) or 69B-Gal4 (7) (Fig. 3 and Movie S1). Embryos were dechorionated in 50% bleach and fixed for 20 min in 4% PFA. GFP and RFP fluorescence was observed without the need for further treatment. Staining of the ventral nerve chord with anti–cleaved caspase 3 (1/50, rabbit, Cell Signaling) was performed as described (5). The embryos depicted in Fig. 4 result from a cross between flies of the following genotypes: UAS-Apoliner5/+; Df(3L)H99/+ and tub-GAL4/+; Df(3L)H99/+. The TUNEL reaction was performed with the Apoptag kit (Qbiogen), according to the manufacturer's instructions. Briefly, embryos were rehydrated from methanol storage, incubated 2–3 h in a working solution of TdT enzyme and the supplied buffer. After washing, embryos were blocked in PBS 4% FCS for 30 min, and incubated overnight in 1/100 dilution of anti-DIG Cy5 antibody (Jackson) in PBS 4% FCS. Embryos were then washed in PBS, Triton 0.1%, mounted in Vectashield (VECTOR Laboratories).
For time-lapse imaging, embryos were dechorionated in 50% bleach for 2 min and mounted in Voltalef oil on a coverslip. Three z-sections (covering 2.6 μm) were collected every 1.5 min. The time-lapse series was assembled and analyzed with Volocity (Improvision). All images were acquired with a Leica SP5 confocal microscope.
Chick Electroporation and Imaging.
Constructs were electroporated into the neural tube of stage HH 10 to 12 (18) chick embryos. After 24 h, embryos were fixed and processed for immunohistochemistry as described previously (17). To reveal Apoliner cleavage (Fig. 5A and C), the following antibodies were used: anti-GFP (1/2000, mouse, Molecular Probes) and anti-RFP (1/1000, rabbit, Chemicon International); secondary: anti-mouse Alexa 488 (1:400, goat, Molecular Probes) and anti-rabbit Alexa 555 (1:400, goat, Molecular Probes). As the caspase antibody was raised in rabbit, it was used on different sections to assess the extent of cell death in the different conditions used (Fig. 5B and D). Antibodies used: anti–cleaved caspase 3 antibody (1/50, rabbit, Cell Signaling) and the secondary anti-rabbit coupled to Cy5 (1/200, goat, Jackson).
Supplementary Material
Acknowledgments.
We thank Eugenia Piddini, Xavier Franch-Marro, Sara Morais da Silva, Liz Hirst, Fabrice Prin, Mark Ditzel, and Francois Leulier for fruitful discussions and technical advice; Cyrille Alexandre for embryo injection, and Liqun Luo for plasmids. This research was funded by the Medical Research Council of Great Britain. The anti-Drosophila actin JL20 developed by J.J.-C. Lin was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806983105/DCSupplemental.
References
- 1.Domingos PM, Steller H. Pathways regulating apoptosis during patterning and development. Curr Opin Genet Dev. 2007;17:294–299. doi: 10.1016/j.gde.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cashio P, Lee TV, Bergmann A. Genetic control of programmed cell death in Drosophila melanogaster. Semin Cell Dev Biol. 2005;16:225–235. doi: 10.1016/j.semcdb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Wang SL, et al. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 4.Ditzel M, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 5.Miguel-Aliaga I, Thor S. Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development. 2004;131:6093–6105. doi: 10.1242/dev.01521. [DOI] [PubMed] [Google Scholar]
- 6.Pazdera TM, Janardhan P, Minden JS. Patterned epidermal cell death in wild-type and segment polarity mutant Drosophila embryos. Development. 1998;125:3427–3436. doi: 10.1242/dev.125.17.3427. [DOI] [PubMed] [Google Scholar]
- 7.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 8.White K, et al. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 9.Clem RJ, Fechheimer M, Miller LK. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 10.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 11.Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 12.Takemoto K, Nagai T, Miyawaki A, Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J Cell Biol. 2003;160:235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuranaga E, Miura M. Nonapoptotic functions of caspases: Caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007;17:135–144. doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenev T, et al. Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 2002;21:5118–5129. doi: 10.1093/emboj/cdf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenev T, et al. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2005;7:70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- 17.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;4:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 18.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.