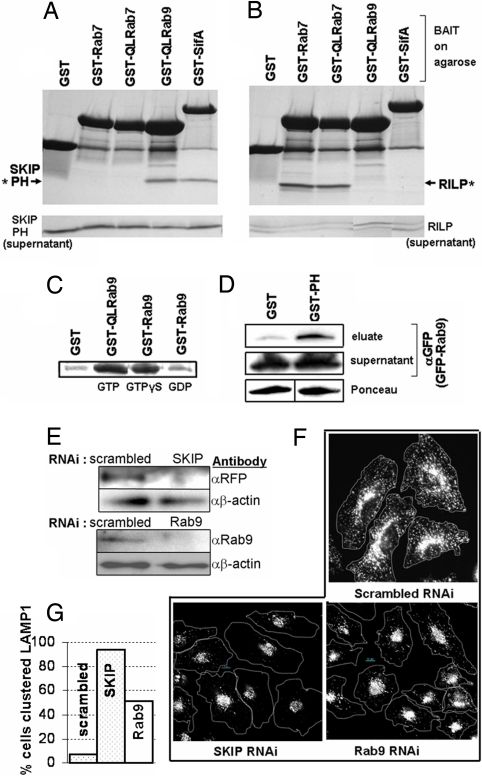
Fig. 1.
Rab9 binds specifically to the PH domain of SKIP, a protein that influences LAMP1 distribution in cells. (A and B) Coomassie stained SDS/PAGE gel shows proteins eluted from washed resin in pulldowns (Upper) or ≈10% of pulldown supernatant (Lower). Resin was loaded with 80 μg of GST, GST-SifA, GST-Q66L-Rab9(GTPγS), GST-Rab7(GTPγS), or GST-Q67L-Rab7(GTP) and incubated with either 53 μg of SKIP PH (amino acids 762–885) (A) or 53 μg of RILP (amino acids 241–320) (B) proteins. Asterisks indicate protein captured and eluted in pulldown. (C) GTP-dependence of binding. SKIP PH from pulldowns is visualized by Coomassie. Resins loaded with 400 μg of GST, GST-Q66L-Rab9, or GST-Rab9 were loaded with the indicated nucleotide and incubated with 400 μg of SKIP PH. (D) SKIP PH can also interact with Rab9 in cell lysates. Resin loaded by using 20 μg of GST or GST-SKIP PH domain was incubated with lysate, created from HeLa cells expressing GFP-Rab9, for pulldown. Approximately 10% of pulldown supernatant is loaded in control blot. (E) Western blots showing knockdown of SKIP or Rab9 protein levels in lysates from cells treated with anti-SKIP, anti-Rab9, or control (scrambled) RNAi. To monitor SKIP knockdown, SKIP RNAi-treated cells were transfected with an RFP-SKIP construct, then lysed and probed by using an anti-RFP antibody. (F) RNAi-treated HeLa cells were fixed, permeabilized, and stained with anti-LAMP1 antibody (white). A dotted line is drawn around cells to indicate the edges. (G) Quantitative analysis of micrographic data, indicating the percentage of cells with clustered LAMP1 in RNAi-treated cells.