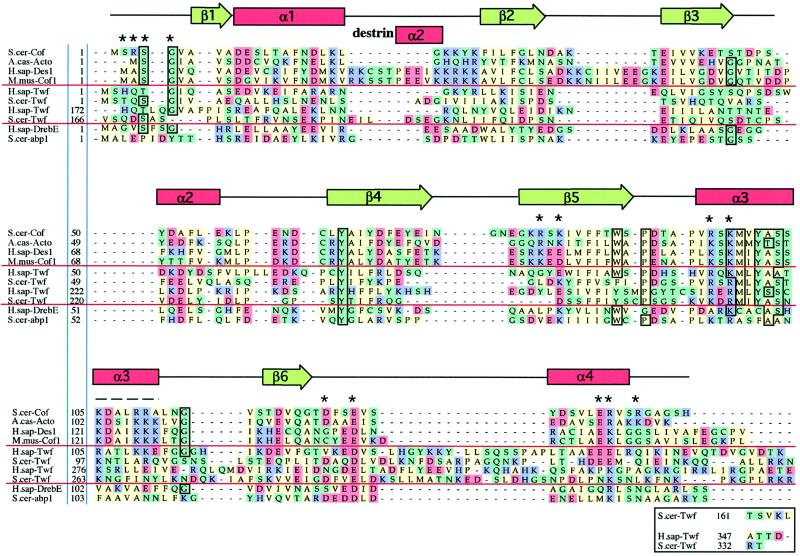
Figure 2.
Sequence alignments of representative examples of ADF-H domains. This alignment was produced essentially according to the methods described for myosin motor domains by Cope et al., (1996). Briefly, the collected sequences were subjected to an initial alignment using the Clustal-W program (Thompson et al., 1994). This initial alignment was refined, thus defining a core ADF-H domain. Acidic (D and E), basic (K, R, and H), uncharged nonpolar (A, I, M, V, L, F, W, and P), and other residues (Y, T, S, G, N, Q, and C) have been colored in red, purple, green, and yellow, respectively. The residues that are >75% conserved throughout the entire ADF-H domain family are boxed. Dashes indicate positions occupied by residues from other ADF/cofilin proteins within the full alignment. The residues shown to be essential for interactions of yeast cofilin with actin (Lappalainen et al., 1997) are indicated by asterisks above the sequences, and the region that has been shown to be important for actin interactions by peptide inhibition studies (Yonezawa et al., 1989) is shown by a dashed line above the sequences. The positions of secondary structure elements based on the yeast cofilin crystal structure (Fedorov et al., 1997) and the nuclear magnetic resonance structure of human destrin (Hatanaka et al., 1996) are shown above the sequences. Protein names, database, and accession numbers for the sequences, respectively, are listed in the legend to Figure 3.