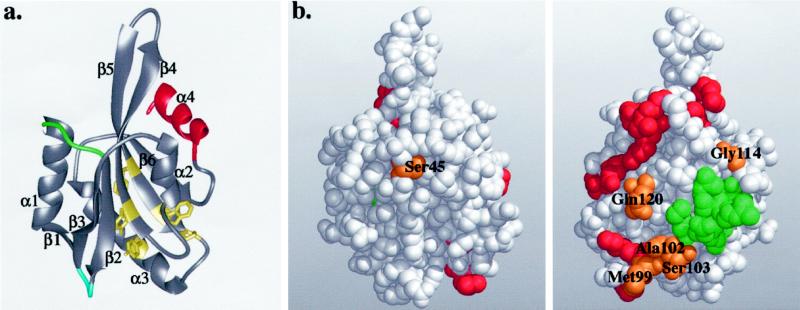
Figure 4.
(a) Ribbon diagram of the yeast cofilin structure. Cofilin has a central mixed β-sheet, which is sandwiched between two pairs of α-helices. The positions of the insertions in mammalian cofilins are in green and blue, and the diverged region in twinfilins is in red. The highly conserved residues that appear to be important for protein stability and correct folding (Tyr64, Phe85, Trp88, Pro90, and Tyr101) are shown in yellow. (b) Space-filling model of yeast cofilin shown in two different orientations (rotated 180° around the y-axis). The residues that are essential for actin interactions in yeast cofilin are highlighted in red. The residues implicated in actin binding by peptide inhibition studies are in green. The highly conserved surface residues (Ser45, Met99, Ala102, Ser103, Gly114, and Gln120) are in orange. All of these residues, except Ser45, are located close to site of cofilin implicated genetically in actin binding, suggesting they also form part of the actin-binding surface. These Figures were produced using Midas Software (University of California San Francisco) running on a Silicon Graphics (Mountain View, CA) Indigo II workstation.