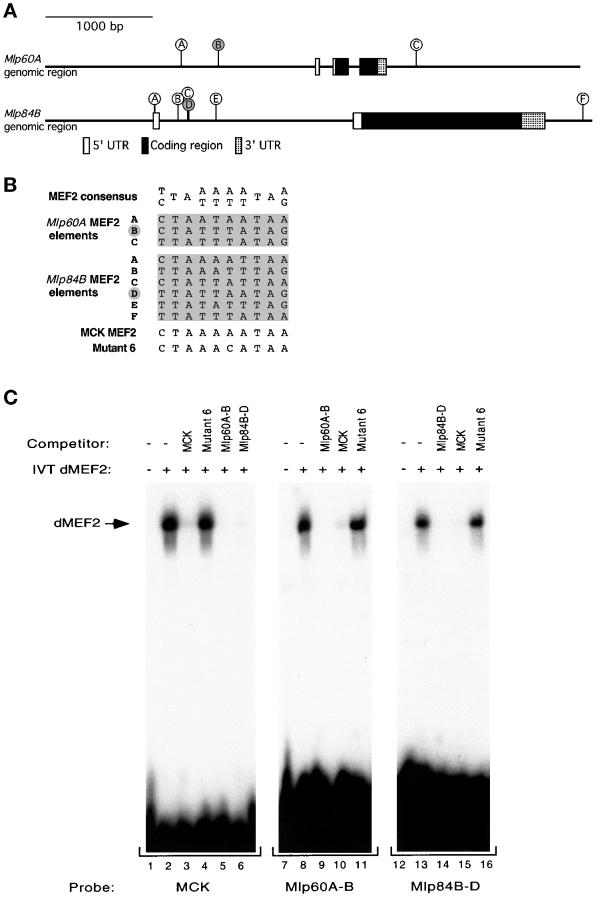
Figure 4.
dMEF2 protein binds to consensus MEF2 sites found in the genomic regions of the Mlp genes. (A) Diagram of the genomic organization of the Mlp60A and Mlp84B genes. The boxed areas indicate the regions that correspond to cDNA sequences. The sticks indicate positions of 10-bp elements that exactly match the MEF2 consensus binding site. The two sites indicated by filled circles are those tested in the in vitro dMEF2 binding assay shown in C. The exact nucleotide range of the Mlp84B sites is noted in GenBank accession number AF090832. (B) The potential MEF2 binding elements found in the putative regulatory regions of Mlp60A and Mlp84B are shown aligned with the MEF2 consensus, the MEF2 regulatory site from the MCK enhancer, and the Mutant 6 form of the MCK site, which does not support MEF2 binding. Letters to the left correspond to the sites diagrammed in A, and the filled letters indicate those sites tested in C. (C) Mobility shift assays with in vitro–translated dMEF2 protein were performed with oligonucleotides corresponding to the MCK MEF2 site and one MEF2 element from each of the Mlp genes. The MEF2 elements used as probes were MCK MEF2 (lanes 1–6), Mlp60A-B (lanes 7–11), and Mlp84B-D (lanes 12–16). For each probe a control lysate lacking translated dMEF2 was included in parallel (lanes 1, 7, and 12). Competitor oligonucleotides were used at 100-fold molar excess of each probe. dMEF2 binds to each of the probes specifically (lanes 2, 8, and 13). Each of the Mlp MEF2 sites binds dMEF2 and competes for binding to the MCK element and their cognate site. A mutant form of the MCK element (Mutant 6) fails to compete for binding with any of the sites tested (lanes 4, 11, and 16).