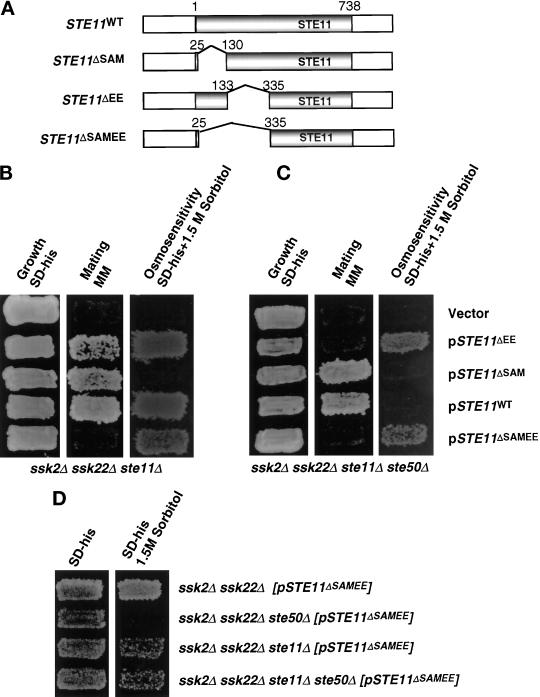
Figure 3.
Requirement for the N-terminal regions of Ste11p in the HOG and the pheromone response pathways. (A) The schematic representation of STE11 and its in-frame deletion constructs (see MATERIALS AND METHODS). The shaded box represents the coding region of STE11, gaps between the shaded boxes represent deletions, and the numbers on two sides of the gap indicate the aa fused after deletion of the gap region. All constructs are low-copy centromeric plasmids. (B) The effect of the N-terminal mutations in a ste11Δ strain. Yeast strain YCW340 (ssk2Δ ssk22Δ ste11Δ) was transformed either with the plasmids indicated or the cloning vector as a negative control, and the transformants were examined for growth on selective medium (left), the ability to form diploids (middle) on minimal medium (MM), and the ability to grow on high-osmolarity medium containing 1.5 M sorbitol (right). (C) The effect of the N-terminal mutations in a ste11Δ ste50Δ strain. Yeast strain YCW555 (ssk2Δ ssk22Δ ste11Δ ste50Δ) was transformed with either the plasmids indicated or the cloning vector as negative control, and examined for growth on selective medium (left), the ability to form diploids (middle) on minimal medium, and the ability to grow on high-osmolarity medium containing 1.5 M sorbitol (right). (D) Wild-type Ste11p has a dominant interfering effect on the constitutively activated allele of Ste11p in the HOG pathway. Yeast strains indicated were transformed with the constitutively active allele of STE11 construct, pSTE11ΔSAMEE, and examined for growth on selective medium (left) and the ability to grow on high-osmolarity medium containing 1.5 M sorbitol (right).