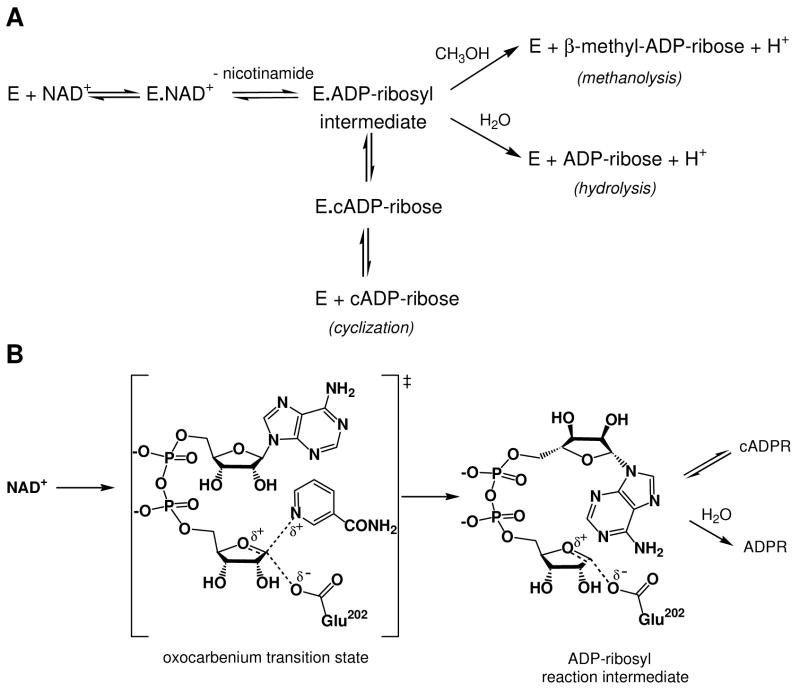
FIGURE 1.
(A) Minimal kinetic mechanism for reactions catalyzed by ADP-ribosyl cyclase family members. (B) Molecular mechanism of the reaction catalyzed by SmNACE. This hypothetical mechanism was adapted from studies on mammalian CD38. The reaction involves the cleavage of the nicotinamide-ribosyl bond via a late transition state leading to the formation of either a covalent acylal ADP-ribosyl intermediate or a carboxylate-stabilized oxocarbenium ion intermediate. In either case the intermediate is then transformed via competing pathways : i) reaction with water yielding ADP-ribose, or ii) intramolecular cyclization to form cyclic ADP-ribose by reaction of N1 of the adenine ring with the C1’’ of the intermediate. The catalytic carboxylate corresponds to Glu202 in SmNACE. At a molecular/structural level, the precise mechanism by which the ‘signature’ Glu124, the second active site carboxylic group, controls the cyclization to hydrolysis ratio remains poorly understood. For a full discussion of the molecular mechanism of these enzymes, see (3).