FIGURE 6.
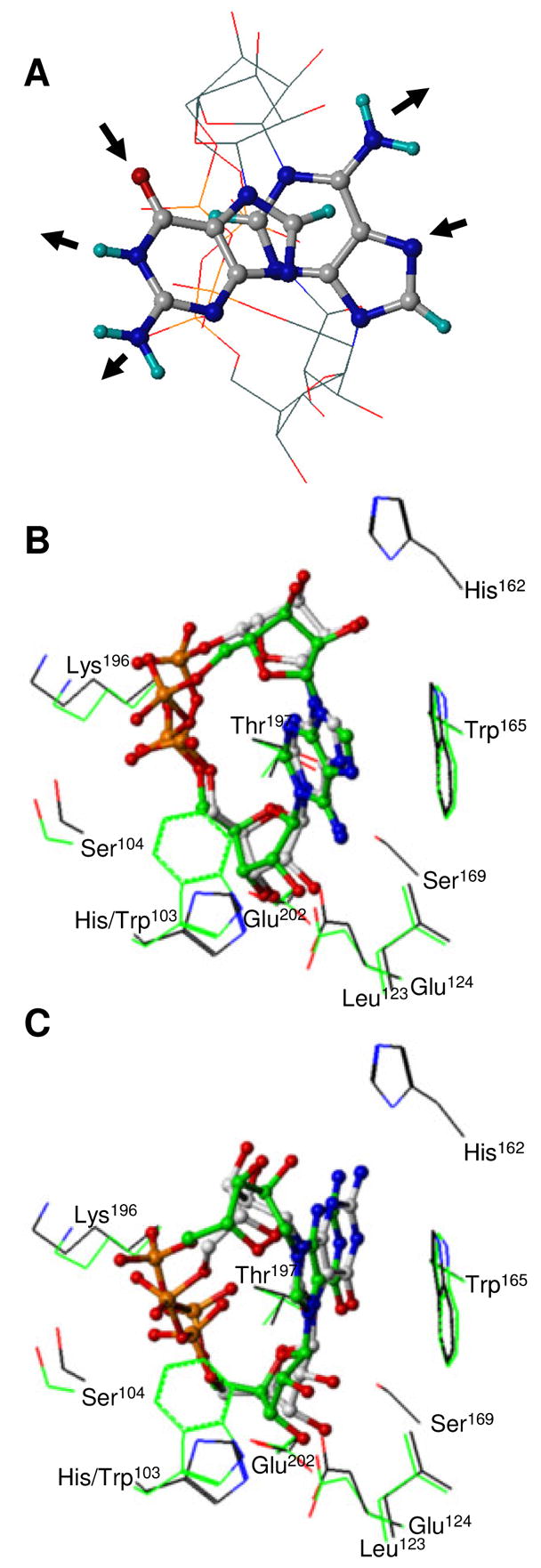
Views of cADPR and cGDPR three-dimensional structure. (A) Free cADPR and cGDPR. Best-fit superposition of the pyrophosphate and ribose heavy atoms of the MacroModel lowest energy conformers. For sake of clarity, ribose and pyrophosphate groups are depicted without hydrogen atoms using line representation whereas all atoms of adenine and guanine bases are shown using ball-and-stick representation. Molecules are colored by atom type as defined in Figure 5. H-bond donors and acceptors in the Watson Crick edge of the bases are indicated using arrows. cADPR (B) and cGDPR (C) docked into the WT and H103W SmNACE active site. Carbon atoms of the WT and mutant complexes are shown in grey and green respectively (the other atoms are colored as defined in Figure 4). All ligand heavy atoms are shown using ball-and-stick. Side chains of the key residues involved in ligand binding are depicted using a line representation and illustrate the surrounding enzyme structure. The rendering was performed using SYBYL 7.1.
