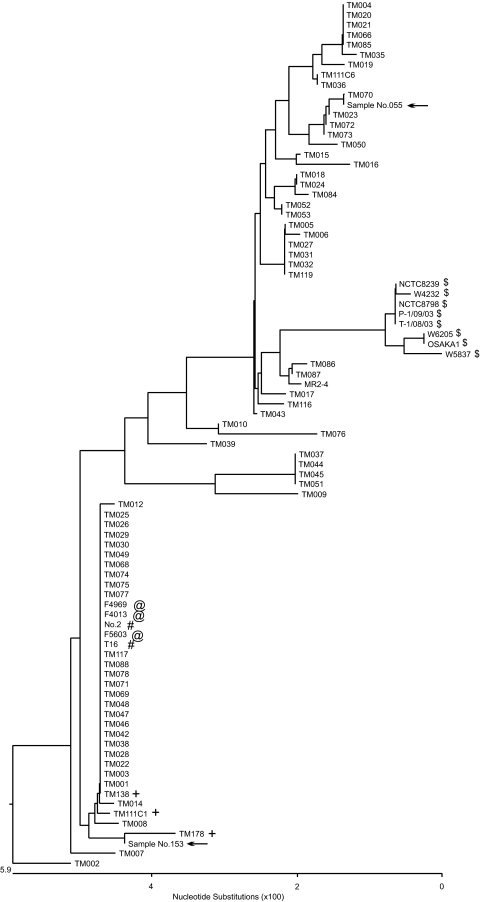
FIG. 2.
Phylogenetic tree analysis of the superoxide gene (sod) of chromosomal cpe strains, plasmid cpe strains, and cpe-negative food isolates. The phylogenetic tree was constructed with 505 bp of sequence information from the sod open reading frame (full length, 760 bp) from food and food poisoning isolates and with 347 bp of sequence information from sod-based assay-positive samples. Symbols: $, chromosomal cpe strains recovered from food poisoning outbreaks in Japan and Europe and chromosomal cpe strains recovered from food isolates in the United States; @, plasmid cpe-positive European diarrhea isolates; #, plasmid cpe-positive strains recovered from food poisoning outbreaks in Japan; +, plasmid cpe-positive food isolates (strain MR2-4 is from a healthy human fecal isolate with cpe on a plasmid; the other strains are cpe-negative food isolates); arrows, sod PCR-positive enriched-culture samples. Chromosomal cpe strains (indicated by the symbol $) formed a distinct cluster which could be easily differentiated from other strains such as plasmid cpe strains. From the sequence information of PCR-positive products from a sod-based assay (indicated with arrows), the PCR products could not have come from a chromosomal cpe strain.