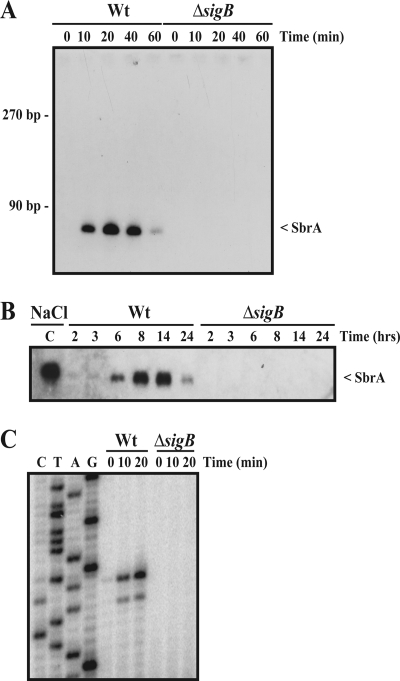
FIG. 1.
Identification of novel σB-regulated sRNAs. (A) Northern blot showing the expression of SbrA after NaCl stress. Overnight cultures of wild-type (Wt) and ΔsigB cells were diluted 100 times in brain heart infusion (BHI) medium and grown to early exponential phase, at which point 4% NaCl was added. The samples were drawn for RNA extraction at the indicated time points after the addition of NaCl. RNA purification and Northern blotting were performed as previously described (6) using a specific 32P-labeled RNA probe prepared by the in vitro transcription of a PCR product generated by using the primers sbrA-Fwd T7 and sbrA-Rev (the primers are listed in Table S1 in the supplemental material). (B) Northern blot showing the expression of SbrA during growth in BHI. Samples were drawn at the indicated time points. The time points of 2, 3, and 6 h correspond to the early to late exponential growth phases, whereas the 8-, 14-, and 24-h time points correspond to the early to late stationary growth phases. Lane C is a positive control corresponding to NaCl-induced SbrA. Wt, wild type. (C) Mapping of the 5′ transcriptional start site of SbrA by primer extension. Wild-type (Wt) and ΔsigB cells were grown in BHI to early exponential phase, at which point 4% NaCl was added. The samples were drawn at the indicated time points. Following RNA extraction, primer extension was performed as previously described (6) using the primer sbrA-1 (see Table S1 in the supplemental material).