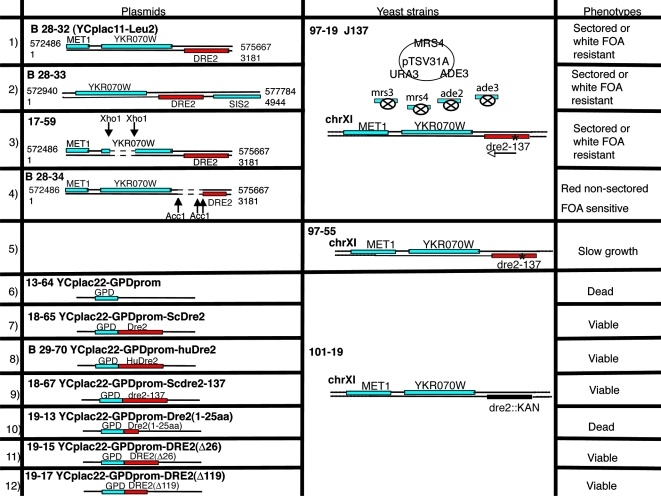
FIG. 2.
Further genetic analysis of synthetic lethal mutant J137. The wild-type copy of the mutated gene in J137 was cloned by complementation with a genomic library in plasmid YCplac111. The transformants formed small nonsectoring red colonies for the most part, with the exception of two single large white colonies. (Rows 1 to 4) The complementing plasmids (rows 1 and 2) were rescued, and complementing activity was further localized by deletions inactivating YKR070W (row 3) or DRE2 (row 4). Only 17-59 retained complementing activity, showing that DRE2 was responsible. (Row 5) The mutation in the genome of J137, called dre2-137, was identified following amplification of the DRE2 open reading frame and flanking regions. A single A-to-T change was present, altering codon 26 from a lysine to a stop codon. The dre2-137 strain, called 97-55, was recovered following backcross of J137 with parental strain 84-9, sporulation, and tetrad dissection. This strain was slow growing but viable and exhibited sensitivity to iron deprivation. (Row 6) A shuffle strain was constructed by heterozygous gene deletion in the diploid, transformation with a Dre2-expressing plasmid with CYH2 marker (18-39), and sporulation. The shuffle strain (101-19) with a Δdre2::kanMX deletion covered by the Dre2 plasmid was completely nonviable following counterselection against the plasmid with cycloheximide. (Row 7) By contrast, the shuffle strain with Dre2 under the control of a strong GPD promoter reinserted on YCplac22 (plasmid 18-65) was viable on cycloheximide. (Row 8) The identical plasmid with the human Dre2 homolog expressed from the GPD promoter (plasmid B29-70) conferred robust growth on the shuffle strain on cycloheximide. (Row 9) The YCplac22-GPD-Dre2 plasmid was mutated, introducing the dre2-137 point mutation (codon 26 changed from lysine to stop), and this plasmid (18-67) was introduced into the shuffle strain, conferring viable growth on cycloheximide. (Row 10) The YCplac22-GPD-Dre2(1-25aa) plasmid (19-13) was transformed into the shuffle strain, resulting in transformants that were not able to grow on a cycloheximide plate. (Rows 11 and 12) However, the plasmids containing N-terminal-truncated Dre2 forms, YCplac22-GPD-Dre2(Δ26) (row 11) and YCplac22-GPD-Dre2(Δ119) (row 12), were able to permit growth on cycloheximide. FOA, fluoroorotic acid.