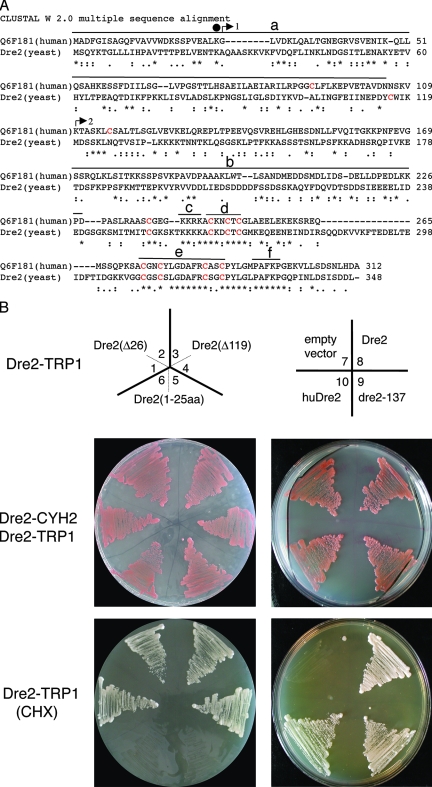
FIG. 3.
Sequence alignment and shuffle strain complementation. (A) Alignment of human Dre2 (Q6F181, also called CIAPIN1 or anamorsin) and yeast Dre2 amino acid sequences using Clustal W 2.0. Underneath the alignment, asterisks represent identical residues, and dots or colons represent lesser degrees of conservative changes. Cysteines are colored red, with eight of nine cysteines perfectly aligned. Above the alignment a black circle over residue 26 indicates the location of the mutation in dre2-137 changing K to stop. Arrows indicate the translational starts of truncated constructs (arrow 1 including positions 27 to 348 labeled Δ26; arrow 2 including positions 120 to 348 labeled Δ119) used in panel B. Overlines indicate possible functional domains: amino-terminal domain (a), acidic and serine-rich patch (b), basic domain (c), CX2CXC domain (d), twin CX2C motifs (e), and region of amino acid identity of unknown significance (f). (B) Complementation of shuffle strain with various Dre2 constructs and human homolog. The shuffle (strain 101-19) with the Δdre::kanMX deletion covered by DRE2 (plasmid 18-39) carrying wild-type DRE2 and CYH2 counterselectable marker was transformed with various Dre2 constructs carried on plasmid YCplac22 and driven by the GPD promoter. The YCplac22 constructs shown in the top panel are as follows: Dre2(Δ26) (1 and 2), Dre2(Δ119) (3 and 4), Dre2(1-25aa) (5 and 6), YCplac22 alone (7), Dre2 open reading frame (8), dre2-137 (9), and human Dre2 open reading frame (10). The transformants with both the counterselectable Dre2-CYH2 and the test Dre2-TRP1 were grown and photographed (middle panel). Then the covering plasmid was removed by plating on cycloheximide (CHX), leaving only the test Dre2-TRP1 plasmid (bottom panel). The results show that DRE2 is required for viability, and the deletion can be rescued by expression of the yeast wild-type protein, the human homolog, the point mutant dre2-137, or the Δ26 and Δ119 truncations but not the fragment with N-terminal amino acids 1 to 25.