FIG. 1.
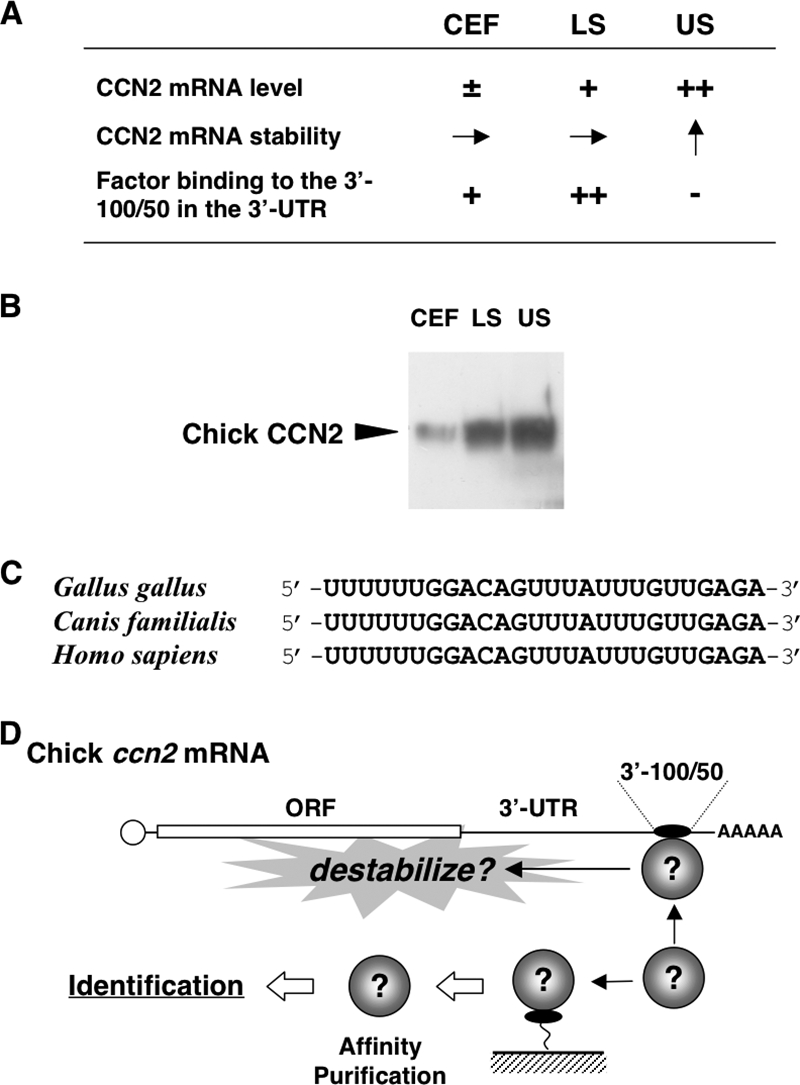
(A) Chicken in vitro model of endochondral ossification and posttranscriptional regulatory outcome of ccn2 expression therein. CEF, LS, and US denote chicken embryonic fibroblasts representing undifferentiated mesenchymal cells, lower sternum cartilage cells representing immature/proliferating chondrocytes, and upper sternum cartilage cells representing hypertrophic chondrocytes, respectively. −, ±, +, and ++ indicate approximate levels of each mRNA or protein in each cell; → and ↑ indicate equivalent and increased mRNA stability, respectively. Note that ccn2 mRNA stability is increased in US cells (arrow pointing up), where binding of a 40-kDa protein to the 3′-100/50 posttranscriptional regulatory element is nearly absent (−). (B) Western blotting analysis of the CCN2 proteins in CEF, LS, and US cells. Total cell lysates comprising 18 μg of DNA were analyzed. (C) A core nucleotide sequence conserved among mammalian species in the 3′-100/50 region and the corresponding sequence in the ccn2 mRNA 3′-UTR. (D) Experimental strategy for the identification of the 40-kDa protein. As summarized in panel A, the protein (illustrated as spheres) binds specifically to the 3′-100/50 RNA fragment in the 3′-UTR, which has been supposed to destabilize ccn2 mRNA. On the basis of this specific RNA-protein interaction, affinity purification methodology was employed, as illustrated. ORF, open reading frame.
