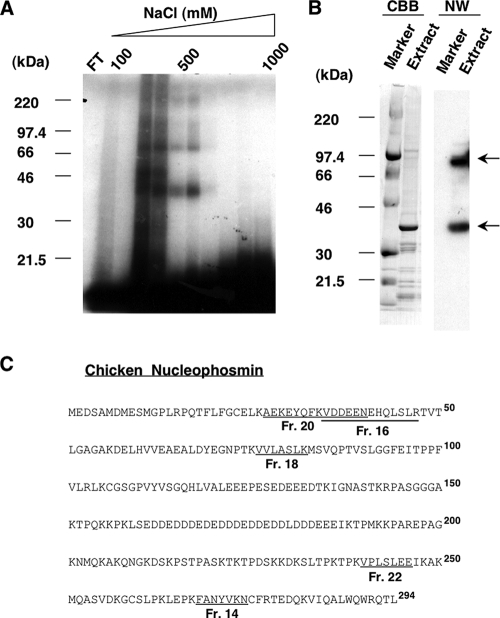
FIG. 2.
Purification and identification of chicken ccn2 mRNA 3′-UTR-binding protein by affinity chromatography. (A) UV cross-linking assay of eluate from a chicken ccn2 mRNA 3′-UTR affinity column. NHS-activated Sepharose in a column was covalently conjugated to the 3′-100/50 fragment of chicken ccn2 mRNA 3′-UTR, which had been transcribed in vitro (see Materials and Methods). The nuclear proteins from CEFs that bound to the RNA were eluted by a linear step gradient of NaCl concentrations (100 to 1,000 mM) and concentrated. The proteins in the column flowthrough (FT) and fractions that bound to the 3′-100/50 RNA fragment were examined by conducting a UV cross-linking assay with SDS-PAGE in a 4 to 20% gradient gel. The positions of molecular mass standards (in kilodaltons) (Rainbow markers; Amersham Bioscience) are shown at the left side of the panel. (B) Northwestern blotting. The molecular mass standards (Marker) and 5 μg of eluted proteins (Extract) from the combined positive fractions (obtained with 200 to 600 mM NaCl in panel A) were subjected to SDS-PAGE in a 4 to 20% gradient gel and blotted onto a PVDF membrane. The membrane was first stained with CBB and then incubated with radiolabeled 3′-100/50 probe for Northwestern analysis (NW). The prominent bands bound by the probe are indicated by arrows at the right side of the panel. (C) Identification of the purified protein as nucleophosmin. A tryptic digestion product of the 40-kDa protein was subjected to Edman degradation, and the internal peptide sequences obtained were analyzed by the BLAST program of the National Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov). Amino acid sequences from five fractions (fractions [Fr.] 14, 16, 18, 20, and 22; see the figure in supplemental material for details) purified through a high-pressure liquid chromatograph column were found to be identical to those of chicken NPM (GenBank accession number NM_205267), as indicated by the fraction numbers and underlined sequences. The boldface superscript numbers to the right of the sequences represent residue numbers of NPM counting from the initiation methionine.