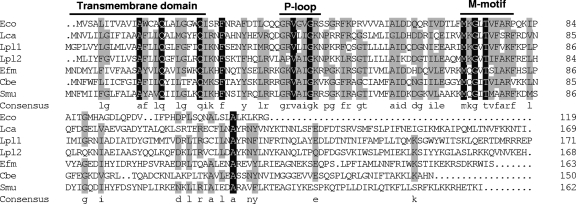
FIG. 3.
Multiple amino acid sequence alignment of GutM from L. casei (Lca, this work), E. coli (Eco, GenBank accession no. NP_417186), L. plantarum (Lpl1; GenBank accession no. NP_786817), L. plantarum (Lpl2; GenBank accession no. NP_786847), Enterococcus faecium (Efm; GenBank accession no. ZP_00604866), C. beijerinckii (Cbe; GenBank accession no. YP_001307479), and S. mutans (Smu, GenBank accession no. AAD33520). The residue number of each protein is indicated on the right. Residues conserved in all sequences are shown against a black background. Residues conserved among at least four of the seven sequences appear against a grey background. The consensus sequence (at least four residues conserved) is shown in lowercase letters. The predicted transmembrane domain and the P-loop motif are shown. M-motif indicates a highly conserved region of unknown function.