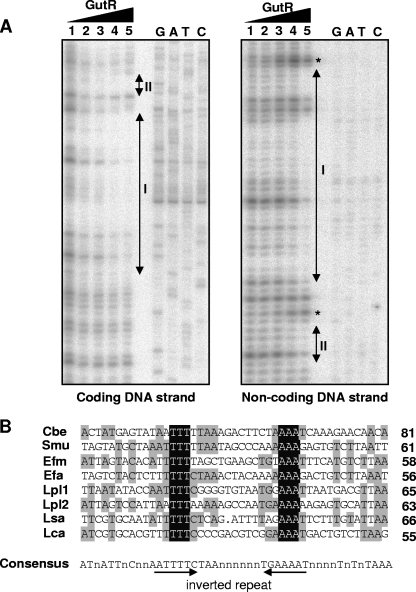
FIG. 4.
(A) DNase I footprinting analysis of GutR binding to the sorbitol promoter region. Protection of the coding and noncoding DNA strands in the presence of 0.00, 0.02, 0.17, 0.68, and 1.36 μg of His-tagged GutR (lanes 1 to 5, respectively). The GutR-protected regions (I and II) in the coding and noncoding DNA strands are indicated by double-arrowhead vertical bars. Asterisks indicate the DNA positions with hypersensitivity to DNase I digestion. Binding positions were determined by the sequence ladder (GATC). (B) Alignment of the GutR-protected region of L. casei (Lca; see panel A) with the hypothetical promoter regions (45 bases) of gut operons from C. beijerinckii (Cbe; GenBank accession no. NC_009617.1), S. mutans (Smu; accession no. AF132127.1), E. faecium (Efm; accession no. NZ_AAAK03000093.1), E. faecalis (Dfa; accession no. NC_004668.1), (Lpl) L. plantarum (accession no. NC_004567.1.1), and L. salivarius (Lsa; accession no. NC_007930.1). At the right end of each sequence lane, we indicated the distance (in bases) to the start codon of the first gene of the corresponding gut operon. Bases conserved in all sequences are shown against a dark background. Bases conserved in at least four of the eight sequences appear against a shaded background. A consensus sequence (50% conservation) is shown below the alignment and contains an inverted repeat marked with arrows.