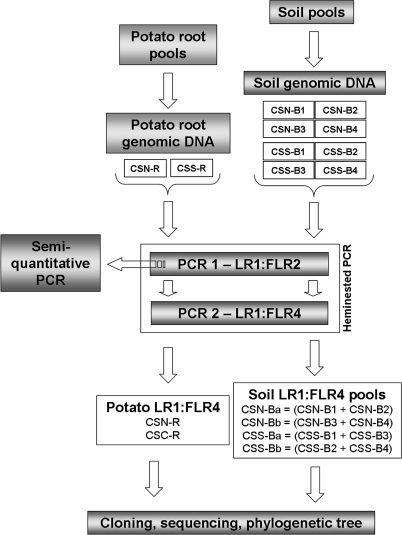
FIG. 1.
Diagram of work flow to prepare the PCR fragment pools used in this study. To prepare PCR fragments from soil, one aliquot of soil for each of the four selected fields per area was used for genomic DNA extraction. Four genomic DNA samples per area (CSN-B1, CSN-B2, CSN-B3, and CSN-B4 and CSS-B1, CSS-B2, CSS-B3, and CSS-B4) were obtained and used in a heminested PCR in which the primer pairs LR1 and FLR2 for the first amplification step and LR1 and FLR4 for the second step were applied. The four LR1/FLR4 heminested PCR products (CSN-B1, CSN-B2, CSN-B3, and CSN-B4 and CSS-B1, CSS-B2, CSS-B3, and CSS-B4) were pooled two by two in equal molar quantities as follows: CSN-B1 and CSN-B2 were pooled to yield CSN-Ba, CSN-B3 and CSN-B4 were pooled to yield CSN-Bb, CSS-B1 and CSS-B3 were pooled to yield CSS-Ba, and CSS-B2 and CSS-B4 were pooled to yield CSS-Bb. The PCR products were then cloned into the pCR4-TOPO vector. To prepare PCR fragments from potato roots, a pooled root sample from each area was obtained by mixing potato roots randomly selected from each field. One root genomic DNA sample (CSN-R or CSS-R) per area was assessed, and one LR1/FLR4 heminested PCR product (CSN-R or CSS-R) per area was cloned into the pCR4-TOPO vector. Finally, inserts from randomly selected clones were sequenced and analyzed by using bioinformatics tools.