Abstract
We investigated the genetic diversity and symbiotic efficiency of 223 Sinorhizobium sp. isolates sampled from a single Mediterranean soil and trapped with four Medicago truncatula lines. DNA molecular polymorphism was estimated by capillary electrophoresis-single-stranded conformation polymorphism and restriction fragment length polymorphism on five loci (IGSNOD, typA, virB11, avhB11, and the 16S rRNA gene). More than 90% of the rhizobia isolated belonged to the Sinorhizobium medicae species (others belonged to Sinorhizobium meliloti), with different proportions of the two species among the four M. truncatula lines. The S. meliloti population was more diverse than that of S. medicae, and significant genetic differentiation among bacterial populations was detected. Single inoculations performed in tubes with each bacterial genotype and each plant line showed significant bacterium-plant line interactions for nodulation and N2 fixation levels. Competition experiments within each species highlighted either strong or weak competition among genotypes within S. medicae and S. meliloti, respectively. Interspecies competition experiments showed S. meliloti to be more competitive than S. medicae for nodulation. Although not highly divergent at a nucleotide level, isolates collected from this single soil sample displayed wide polymorphism for both nodulation and N2 fixation. Each M. truncatula line might influence Sinorhizobium soil population diversity differently via its symbiotic preferences. Our data suggested that the two species did not evolve similarly, with S. meliloti showing polymorphism and variable selective pressures and S. medicae showing traces of a recent demographic expansion. Strain effectiveness might have played a role in the species and genotype proportions, but in conjunction with strain adaptation to environmental factors.
The rhizobium-legume nitrogen-fixing association is a good model for studying symbiosis and coevolution between organisms. In this mutualistic association, bacteria form nodules on plant roots (more rarely on the stem), where atmospheric nitrogen is reduced to ammonium available for the plant. Among the various plant-bacterium couples studied so far, the Medicago truncatula association with Sinorhizobium meliloti is particularly interesting (8) and has often been studied as a model system for genetic description of the molecular pathways involved in the establishment of the symbiosis (16). M. truncatula has the ability to form an efficient symbiosis with two bacterial species, S. meliloti (9) and its sister species, Sinorhizobium medicae (29).
The genetic diversity of these two bacterial species, especially S. meliloti, has usually been described as quite important (2, 7, 11, 40). This bacterial diversity, assessed by phenotypic or genotypic analyses, has been reported to be influenced by several factors, including geographical location (32), soil factors (14), and plant species (19, 37). Furthermore, at an interspecies level, several studies on S. meliloti and S. medicae natural populations, trapped from various soils (using several Medicago species), reported either the balanced coexistence of the two bacterial species (3, 23) or the dominance of one of them (3, 31, 33, 43).
At a plant genotype level, host nodulation specificity could also influence the bacterial diversity in soil. Significant variations were detected among rhizobial populations associated with different plant lines of Vicia faba (38), Pisum sativum (10), or Medicago sativa (26). Symbiotic specificity studies among M. truncatula lines were based on single-strain inoculations. Some of them showed qualitative symbiotic polymorphism for nodulation (Nod+/Nod−) and nitrogen fixation (Fix+/Fix−) among a wide collection of M. truncatula fixed lines inoculated with diverse S. meliloti strains (35, 39). Other studies demonstrated that different plant-rhizobium combinations displayed different quantitative levels of effectiveness, described as the input that a bacterial nitrogen-fixing symbiosis makes to plant nitrogen metabolism, with S. meliloti (27) and S. medicae species (17). However, as a host plant usually interacts with several symbionts, these studies do not reflect possible quantitative variations of symbiotic effectiveness within a bacterial population encompassing diverse genotypes. Studying plant symbiotic specificity when put in contact with a single soil would reflect what occurs in a natural ecosystem compared to artificial single inoculations.
Nodulation competition among S. meliloti and S. medicae genotypes, at both intra- and interspecies levels, is another biotic factor which could also influence the genetic structure of bacterial populations recovered from soil. Strain competitiveness (defined as the ability to dominate nodulation) experiments were mostly performed to compare inoculated strains versus native bacteria, in order to enhance the agricultural yield by selecting the best strains for nodulation and nitrogen fixation efficiency (21, 42).
In the M. truncatula-Sinorhizobium symbiotic system, it is still unclear (3) if the plant cultivar and competition among bacterial genotypes acting at both inter- and intraspecies levels are true selective forces driving the partner choice in nitrogen fixation association. Furthermore, a study of the nitrogen fixation polymorphism of these bacteria within a single geographical site has never been performed. With the aim of improving our understanding of the bacterial population structure and diversity maintenance in this symbiotic association, we carried out a study whose objectives were (i) to explore the genetic diversity of symbiotic bacteria recovered from a single soil and trapped with four different M. truncatula lines, (ii) to test for significant symbiotic divergence among Sinorhizobium populations sampled with four different M. truncatula lines, (iii) to perform competition assays within and between the two bacterial species S. meliloti and S. medicae to evaluate strain competitiveness, and (iv) to compare the fixation efficiencies of single-strain inoculants and multistrain mixtures.
MATERIALS AND METHODS
Bacterial trapping from soil samples.
Soil samples were collected in an old abandoned vineyard fallow parcel at Saint Bauzille de la Sylve, France (43°37′N, 3°33′E). The soil was characterized as loamed limestone with a pH of 8.3. Soil samples were taken at five points (at a 2-m distance from each other) in the plot (depth, 5 to 25 cm) and were homogenized in the laboratory in a single bulk batch.
Aliquots of soil were put into contact with four M. truncatula fixed lines, originally sampled from different countries: (i) DZA315-16 (Algeria), (ii) F83-005 (France), (iii) PRT180-21 (Portugal), and (iv) the reference line, JemalongA17. These lines were chosen based on the data of Ronfort et al. (30), which showed large genetic distances among them using microsatellite data. After seed sterilization, five individuals per line were cultivated for 2 months on a soil aliquot, and nodules were collected from plant roots and stored at −80°C in 20% glycerol till they were used. From 9 to 14 nodules per plant were surface sterilized, and isolation of a single bacterial clone per nodule was performed.
DNA extraction and marker choice for genetic-diversity analyses.
Bacteria were grown in 20 ml yeast extract-mannitol liquid medium for at least 3 days at 27°C under agitation (41), and total genomic DNA was isolated (3).
The genome of S. meliloti Sm1021 comprises one chromosome (3.65 Mb) and two megaplasmids (pSymA and pSymB), whereas that of S. medicae strain WSM419 comprises in addition one small plasmid. We amplified five genomic regions evenly distributed along the genome and implicated in different bacterial functions. Markers are located either within genes or adjacent to them within an intergenic spacer (IGS). We used two markers involved in symbiosis: IGSNOD (on pSymA), framed by nodulation genes involved in symbiosis (nodE and nodG), and typA (on the chromosome), which is essential for symbiosis on M. truncatula JemalongA17 (20). We also used two paralogous genes, virB11 and avhB11, implicated in effector translocation and conjugation, respectively, in Agrobacterium tumefaciens. In this study, we named loci virB11 and avhB11 as described in A. tumefaciens. In the S. meliloti reference strain (Sm2011), a single gene, named virB11, has been found (on pSymA) with high similarity to avhB11 of A. tumefaciens. On the other hand, the S. medicae reference strain (WSM419) also contains a single locus, also named virB11, but with high similarity to virB11 of A. tumefaciens. Furthermore, we studied the 16S rRNA gene, which allows species discrimination between S. meliloti and S. medicae.
PCR-CE-SSCP analyses.
Allelic discrimination was performed by capillary electrophoresis-single-stranded conformation polymorphism (CE-SSCP) (36). For the PCRs used for the SSCP, the primers of each set were 5′ labeled with two different fluorescent dyes, FAM (5-carboxyfluorescein) and HEX (5-carboxy-4′,5′,2′,4′,5′,7′-hexachlorofluorescein) (Table 1), and we used PCR mixtures and cycling conditions without a touchdown program, as described previously (3). Precipitation of the PCR products was performed by adding 1/10 sodium acetate volume (sodium acetate-EDTA buffer [1.5 M sodium acetate, pH >8, and 250 mM EDTA]) and 4 volumes of 95% ethanol and centrifuging the mixture at 4,000 rpm and 10°C for 35 min. The pellet was washed in 100 μl 70% ethanol and then centrifuged again for 15 min. The DNA was resuspended in 50 μl sterile Tris-HCl (10 mM).
TABLE 1.
Primers used in the study
| Locus | Primers | Sequence (5′-3′) | Tma (°C) | PCR product size (bp) |
|---|---|---|---|---|
| IGSNOD | nod283R_HEX | GCGATCCATTCACGCTGAT | 55 | 304 |
| nod717F_FAM | CAGTTCTGGCATTCAAGC | |||
| typA | typ1706R_HEX | GGGCTGGTAGTCGTGGAACA | 59 | 258 |
| typ1707F_FAM | GCACAAGGATGAGAACGGACAG | |||
| virB11 and avhB11 | virB1478F_FAM | SGGTWSSGGCAAGACGAC | 56 | 212 |
| virB1479R_HEX | AGCARWATTCGGTCCGGC | |||
| virB11 | virB11F_383 | GAGCGGCTACAGATCTGTT | 53b | 492 |
| virB11R_860 | CRTCGCGYATCTCGCCGAGC | |||
| avhB11 | avhB11F_383 | GARCGVATCCAGATCGTCA | 53b | 483 |
| avhB11R_860 | CRTCKCGMARYTCCTGMAGC | |||
| 16S rRNA gene | FGPS 1509 | AAGGAGGGGATCCAGCCGCA | 57 | 1,500 |
| FGPS 6 | GGAGAGTTAGATCTTGGCTCAG |
Tm, melting temperature.
A touchdown PCR program was used (20 cycles of annealing at 63°C [decreasing by 0.5°C at each cycle] followed by 25 cycles of annealing at 53°C).
CE-SSCP analyses of PCR products were performed on a MegaBACE 1000 DNA Analysis System (Amersham Biosciences) equipped with 96 coated capillaries using 70-min runs at 40°C and 8 kV. The raw data were exported in RSD format, different runs were aligned, and peaks were detected with MegaBACE Genetic Profiler V1.5 software (Amersham).
Nucleotide sequencing and PCR-RFLP analyses of the 16S rRNA gene.
We sequenced each different CE-SSCP allele obtained. For similar alleles, we randomly selected and sequenced from 1 to 26 individuals each. PCR amplifications were performed as described above using unlabeled primers, except for virB11 and avhB11 (Table 1). Bands were extracted from 1% agarose gels and purified using the Qiaquick PCR purification kit (Qiagen). Sequencing reactions were performed by the Macrogen Company. All isolates were characterized by PCR-restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA gene (Table 1) with the restriction enzyme RsaI, which differentiates the two Sinorhizobium species.
Single- and mixed-inoculant preparation.
We tested each of the 13 SSCP genotypes isolated from soil for effectiveness with the four plant lines and for competition for nodulation (using mixed-strain inoculants). We estimated the correlation between the optical density at 620 nm and cell numbers (determined by plate counts) for the 13 reference strains separately (data not shown). In the first step, we carried out single-strain inoculations. In the second step, two bacterial mixtures containing, respectively, the nine genotypes of S. meliloti and the four genotypes of S. medicae were prepared. In the third step, an interspecies bacterial competition was performed by mixing the dominant genotype for nodulation of each bacterial species within each plant line, according to results obtained from the intraspecies competition. Mixed inoculants were prepared by combining equal quantities (i.e., the same volume at the same optical density at 620 nm) of each single-strain inoculant. In all experiments, the inoculum was adjusted to approximately 5 × 106 cells ml−1.
Plant growth and data collection in the competition and single-strain inoculant experiments.
Plants were individually grown in tubes (215 by 20 mm) containing 20 ml of inclined sterile nutritive medium (15 g liter−1 of agar) and 50 ml of liquid sterile nutritive solution diluted four times. After sterilization and germination, each seedling was put into a tube and inoculated with 1.5 ml inoculum 2 days later. Negative control plants received only sterile water, and positive controls received 5 mM nitrogen [Ca(NO3)2]. The plants were grown for 7 weeks with a 16-h photoperiod (alternating 24°C and 16°C). Ten replicates were used per plant line and treatment. The total number of nodules and the aerial-shoot biomass on all plants of each experiment were obtained. We estimated plant fitness by the dry aerial biomass, which is strongly correlated with the fixation efficiency (18, 24, 39). Nodules were harvested in the competition plant assays, and one bacterial isolate was purified from each nodule. For the intraspecific-competition assay, bacterial isolates were genotyped as described above by PCR sequencing on IGSNOD for S. medicae and on the three loci IGSNOD, typA, and virB11 for S. meliloti, as these markers were sufficient to distinguish each genotype. In the interspecific-competition experiment, PCR-RFLP on the 16S rRNA gene was used for genotyping bacterial species.
Statistical analysis of genetic diversity and population differentiation.
Diversity indexes of S. meliloti and S. medicae bacterial populations obtained from trapping were calculated at each locus using two summary statistics, allelic richness and Nei's diversity index, using Genclone 2.0 (1). The number of polymorphic sites (S), the number of analyzed sites, the estimator of nucleotide diversity, and Watterson's estimate of the scaled mutation rate were computed at each locus. When applicable (i.e., S > 0), Tajima's D statistic was calculated and departure from the neutral evolutionary model was tested for each species at each locus using Arlequin 3.1 (12).
The genetic differentiation among bacterial populations isolated from each plant line was tested by exact tests of global genotypic differentiation using Genepop 3.4 (28). Significant departures from equal proportions of the two species in both trapping and interspecific-competition experiments were tested with a chi-square test using Statistica 6.
Plant fitness and nodulation ability analyses.
Based on the single-inoculation experiment results, we tested the effects of plant lines, bacterial species, bacterial genotypes nested within species, and the two plant line-bacterial species and plant line-bacterial genotype interactions using an analysis of variance performed with JMP 7.0.1 (SAS Institute Inc.). Based on the two intraspecific competitions (the strain mixture for each species), we used an analysis of variance to test the effects of plant lines, bacterial species, and their interaction. Least-significant-difference Fisher post hoc tests were performed to compare the plant biomasses induced by the mixtures to those of the single-strain inocula.
Nucleotide sequence accession numbers.
All sequences have been deposited in the GenBank database under the following accession numbers: IGSNOD, AM991101 to AM991108; typA, AM991109 to AM991113; virB11, AM991116, AM992458, AM992459, and AM993155; and avhB11, AM991114 and AM991115.
RESULTS
Bacterial diversity.
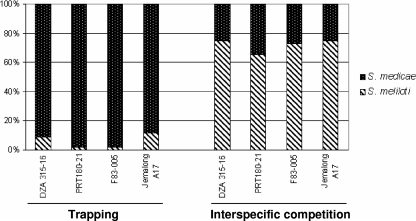
A total of 225 Sinorhizobium isolates were obtained, with 52 to 59 individuals from each of the four M. truncatula lines. The proportion of S. medicae was significantly higher than the proportion of S. meliloti (Fig. 1) (χ2 = 169.27; df = 7; P < 0.001).
FIG. 1.
Compositions of the bacterial populations in trapping and in the interspecific competition experiment.
Bacterial genotyping by PCR-CE-SSCP yielded from one to four different alleles per bacterial species and locus (Table 2). From 10 to 37 isolates per locus were sequenced to confirm discrimination among alleles, and a nearly perfect correspondence was found between the two methods. Only one new allele was detected when sequencing for virB11, with alleles E and W giving the same SSCP profile. All alleles were specific to a bacterial species.
TABLE 2.
Genotypes of S. medicae and S. meliloti isolates from soil trappinga
| Species | Multilocus genotype | No. of individuals in the soil trapping | Reference isolate | Allele at each locus
|
No. of sequenced isolates
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S | IGSNOD | typA | virB11 | avhB11 | IGSNOD | typA | virB11 | avhB11 | ||||
| S. medicae | md1 | 202 | STM 5484 | E | J | P | W | NAb | 6 | 6 | 23 | |
| md2 | 2 | STM 5465 | E | O | P | W | NA | 2 | ||||
| md3 | 3 | STM 5474 | E | K | P | W | NA | 3 | 2 | |||
| md4 | 2 | STM 5477 | E | T | P | W | NA | 2 | 1 | |||
| S. meliloti | ml1 | 2 | STM 5480 | I | B | A | Q | D | 2 | 2 | 2 | 2 |
| ml2 | 3 | STM 5457 | I | R | M | H | D | 3 | 3 | 2 | 2 | |
| ml3 | 1 | STM 5482 | I | V | C | NA | N | 1 | 1 | 1 | ||
| ml4 | 1 | STM 5466 | I | V | A | Q | NA | 1 | 1 | 1 | ||
| ml5 | 1 | STM 5463 | I | V | M | E | NA | 1 | 1 | 1 | ||
| ml6 | 1 | STM 5473 | I | V | U | Q | D | 1 | 1 | 1 | 1 | |
| ml7 | 4 | STM 5472 | I | V | A | H | N | 4 | 4 | 2 | 2 | |
| ml8 | 1 | STM 5478 | I | L | U | H | D | 1 | 1 | 1 | 1 | |
| ml9 | 1 | STM 5481 | I | B | U | H | D | 1 | 1 | 1 | 1 | |
Total numbers are as follows: genotypes, 13; individuals, 224; alleles, 2 (16S), 8, (IGSNOD), 5 (typA), 5 (virB11), and 3 (avhB11).
NA, not amplified. Allele names were given randomly.
As in the sequenced S. medicae strain, we detected only virB11 in each S. medicae isolate. Within S. meliloti strains, we detected either two genes (avhB11 and virB11), only the virB11 gene (as in the sequenced S. medicae strain, WSM419), or only the avhB11 gene (as in the sequenced S. meliloti strain, Sm2011).
Using all loci, 13 different multilocus genotypes were detected, four and nine, respectively, for S. medicae and S. meliloti (Table 2). At least for the bacterial populations isolated from plant lines F83-005 and PRT180-21, S. meliloti diversity appeared larger than that of S. medicae in terms of allelic richness and genotypic distribution. Nucleotide divergence among strains was low for each locus, especially for S. medicae (Table 3). Tajima's D value for the different loci of S. meliloti did not depart from a neutral hypothesis of evolution. In S. medicae, the most negative D value was found for IGSNOD, deviating significantly from neutrality.
TABLE 3.
Genetic diversity of S. medicae and S. meliloti populations from soil trapping
| Locus | No. of samplesa | Genotypic diversityb
|
Nucleotide diversityc
|
|||||
|---|---|---|---|---|---|---|---|---|
| R | H | S | L | π | θw | Tajima's D | ||
| S. medicae | 209 | |||||||
| IGSNOD | 4 | 0.18 | 4 | 253 | 0.0004 | 0.0527 | −1.2951d | |
| typA | 1 | 0 | 0 | 227 | 0 | 0 | NC | |
| virB11 | 1 | 0 | 0 | 167 | 0 | 0 | NC | |
| avhB11 | 1 | 0 | 0 | 0 | 0 | 0 | NC | |
| Multilocus | 4 | 0.18 | ||||||
| S. meliloti | 15 | |||||||
| IGSNOD | 4 | 1.16 | 6 | 253 | 0.0056 | 1.5887 | −1.0095 | |
| typA | 4 | 1.40 | 5 | 227 | 0.0086 | 1.9189 | 0.8887 | |
| virB11 | 4 | 0.95 | 2 | 167 | 0.0035 | 0.8728 | 0.2006 | |
| avhB11 | 3 | 0.97 | 1 | 213 | 0.0024 | 0.7863 | 1.3004 | |
| Multilocus | 9 | 2.03 | ||||||
| Sinorhizobium sp. | 224 | |||||||
| Multilocus | 13 | 0.55 | ||||||
Number of isolates.
R, allelic richness; H, Nei's diversity index.
S, number of polymorphic sites; L, number of analyzed sites; π, the estimator of nucleotidic diversity; θw, Watterson's estimate of the scaled mutation rate; NC, not calculable.
P ≤ 0.05.
Among the four genotypes found within S. medicae species, the genotype md1 was dominant, representing more than 92% of the isolates in the four plant line populations (Table 4). For the DZA315-16 and JemalongA17 plant populations, for which more than one S. meliloti strain was recovered, the proportion of each S. meliloti genotype ranged from 12.5 to 40%.
TABLE 4.
Percentages of genotypes of S. medicae and S. meliloti isolates from trapping and intraspecific competitions in four plant lines
| Genotype | %a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Soil trapping
|
Intraspecific competition
|
|||||||
| DZA315-16 | PRT180-21 | F83-005 | JemalongA17 | DZA315-16 | PRT180-21 | F83-005 | JemalongA17 | |
| S. medicae | ||||||||
| md1 | 100 | 96.4 | 98 | 92 | 0 | 0 | 0 | 0 |
| md2 | 0 | 3.6 | 0 | 0 | 3.3 | 11.1 | 3.3 | 2.4 |
| md3 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 |
| md4 | 0 | 0 | 2 | 2 | 96.7 | 88.9 | 96.7 | 97.6 |
| S. meliloti | ||||||||
| ml1 | 20 | 0 | 0 | 12.5 | 19.4 | 6.1 | 3.1 | 5.9 |
| ml2 | 20 | 0 | 100 | 12.5 | 0 | 0 | 0 | 0 |
| ml3 | 0 | 0 | 0 | 12.5 | 19.4 | 24.2 | 6.3 | 11.8 |
| ml4 | 0 | 100 | 0 | 0 | 25.8 | 12.1 | 3.1 | 23.5 |
| ml5 | 20 | 0 | 0 | 0 | 25.8 | 30.3 | 15.6 | 11.8 |
| ml6 | 0 | 0 | 0 | 12.5 | 3.2 | 15.2 | 0 | 2.9 |
| ml7 | 40 | 0 | 0 | 25 | 0 | 3.2 | 3.1 | 0 |
| ml8 | 0 | 0 | 0 | 12.5 | 6.5 | 9.1 | 68.7 | 44.1 |
| ml9 | 0 | 0 | 0 | 12.5 | 0 | 0 | 0 | 0 |
The numbers in boldface indicate the highest proportion found for each bacterial species-plant line combination in the two experiments. The numbers of isolates were as follows: S. medicae, 51 (DZA315-16, F83-005, and JemalongA17) and 56 (PRT180-21) for soil trapping and 36 (DZA315-1), 30 (PRT180-21 and F83-005), and 42 (JemalongA17) for intraspecific competition and S. meliloti, 5 (DZA315-16), 1 (PRT180-21 and F83-005), and 8 (JemalongA17) for soil trapping and 31 (DZA315-16), 33 (PRT180-21), 32 (F83-005), and 34 (JemalongA17) for intraspecific competition.
Bacterial population differentiation among plant lines.
The overall genotypic differentiation among the four S. medicae populations was barely significant (G-like test, P = 0.02). Conversely, we could not detect any significant differentiation among S. meliloti populations (G-like test, P = 0.62), which might result from the very small sample size of each population. Furthermore, the proportions of S. medicae and S. meliloti differed among the four plant lines, but not strongly (G-like test, P = 0.04).
Intraspecific competition with all species genotypes.
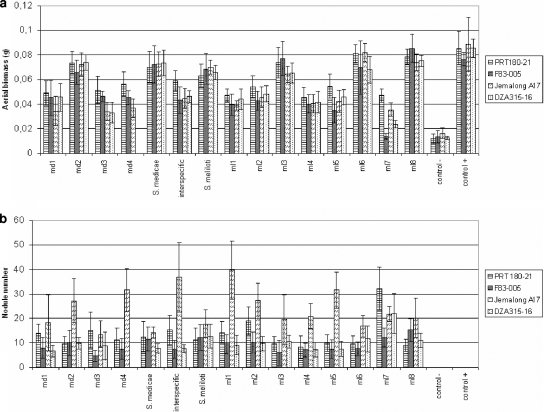
Each of the 13 different bacterial genotypes was tested independently for nodulation. Only the S. meliloti ml9 strain could not renodulate any of the four plant lines (Fig. 2). This strain might be an opportunist rhizobium (coinfecting a nodule), which might not harbor all the symbiotic genes.
FIG. 2.
Mean dry aerial biomass (± standard error) (a) and mean nodule number (± standard error) (b) obtained for each plant line-bacterial genotype combination. The four S. medicae (md1 to md4) and the eight S. meliloti (ml1 to ml8) strains were inoculated separately on each M. truncatula line. “S. medicae” and “S. meliloti” indicate the results of the two mixtures including all the strains for each species, and “interspecific” is the result of a mixture including one strain of each species (see the text). “control −” is the negative control (no inoculation), and “control +” is the positive control (5 mM nitrogen).
When the four S. medicae genotypes were inoculated on each plant line in an equal mixture, only two genotypes (md2 and md4) were reisolated from the 138 nodules genotyped (Table 4), with genotype md4 being significantly dominant (χ2 = 363.48; df = 15; P < 0.001). It is noteworthy that, conversely, the most frequent genotype, md4, had been recovered at a very low frequency in the trapping experiment.
Seven of the nine different S. meliloti genotypes isolated from the soil-trapping experiment were recovered from the 130 nodules genotyped after plant inoculation with an equal mixture of the nine strains (Table 4). Two genotypes (ml2 and ml9) were not reisolated in this experiment. The chi-square tests could not rule out the hypothesis of equal frequencies among strain genotypes in DZA315-16 (χ2 = 8.69; df = 5; P = 0.12), whereas one genotype was found significantly more frequently in other plant lines (data not shown). Finally, the four S. meliloti populations showed highly significant genotypic differentiation (G-like test, P < 0.001).
Interspecific competition with two species genotypes.
Genotype md4 for S. medicae was systematically used and mixed with either S. meliloti genotype ml8 (for the F83-005 and JemalongA17 lines) or ml5 (for the PRT180-21 and DZA315-16 lines). One hundred and twenty-one nodules were genotyped. S. meliloti strains were recovered significantly more frequently than S. medicae strains in each of the four populations (χ2 = 23.99; df = 7; P < 0.001) (Fig. 1).
Plant fitness statistical analyses.
Each plant line was inoculated with each single bacterial genotype isolated, with each intraspecific mixture, or with the mixture of the two dominant strains of each bacterial species. The means and variances of the aerial biomasses and nodule numbers are shown in Fig. 2. Measurements of DZA315-16 plants inoculated with genotype md4 are absent, due to technical problems.
(i) Single inoculants.
Direct estimation of plant line-bacterial genotype (species) interaction was not possible due to the nonequilibrated data we obtained because of the absence of the combination DZA315-16/md4. Therefore, we included in our data virtual data corresponding to this missing combination. We tested several virtual conservative measures (of the nodule number and aerial biomass), i.e., the mean of the md4 strain, the mean of the DZA315-16 line, and the mean of all S. medicae strains. The final results were independent of the choice of the mean (data not shown), suggesting the reliability of such an approach to compensate for missing data.
For both the dry aerial biomass and nodule number measures of all single genotypes, the plant line, bacterial species, bacterial genotype (species), and plant line genotype (species) interaction effects were highly significant. Plant line-bacterial species interaction was found to be barely significant for the aerial biomass and not significant for the nodule number (Table 5, all single genotypes).
TABLE 5.
Analyses of variance for dry aerial biomasses and nodule numbers of plants
| Inoculanta | Effect | df | Aerial biomass
|
Nodule no.
|
||
|---|---|---|---|---|---|---|
| F | Pb | F | Pb | |||
| All single genotypes | Plant line | 3 | 24.8 | *** | 118.6 | *** |
| Bacterial genotype (species) | 10 | 120.8 | *** | 10.6 | *** | |
| Bacterial species | 1 | 7.3 | ** | 11.1 | *** | |
| Plant line-bacterial species | 3 | 2.9 | * | 1.0 | NS | |
| Plant line-bacterial genotype (species) | 30 | 3.8 | *** | 5.3 | *** | |
| Two intraspecific mixtures | Plant line | 3 | 0.8 | NS | 5.7 | ** |
| Species mixture | 1 | 5.1 | * | 2.9 | NS | |
| Plant-bacteria | 3 | 0.2 | NS | 1.8 | NS | |
| S. medicae (single | Plant line | 3 | 10.0 | *** | 52.1 | *** |
| genotypes and mixture) | Bacterial genotype | 4 | 102.3 | *** | 2.8 | * |
| Plant-bacteria | 12 | 2.6 | ** | 4.2 | *** | |
| S. meliloti (single | Plant line | 3 | 15.2 | *** | 100.1 | *** |
| genotypes and mixture) | Bacterial genotype | 8 | 137.6 | *** | 13.6 | *** |
| Plant-bacteria | 24 | 4.7 | *** | 6.2 | *** | |
The plants were inoculated with (top to bottom) the 12 bacterial genotypes independently, the two intraspecific mixtures, the four S. medicae genotypes and the S. medicae intraspecific mixture, or the eight S. meliloti genotypes and the S. meliloti intraspecific mixture.
*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; NS, P > 0.05.
Post hoc tests were performed to rank the various parameters within each effect studied: S. meliloti induced a greater dry biomass than S. medicae (P = 0.02) on each of the four plant lines. Within S. medicae, genotype md2 induced a better biomass than the three other S. medicae genotypes (P < 0.005), while the eight S. meliloti genotypes were grouped in five clusters differing from each other significantly in their nitrogen fixation levels (data not shown).
S. meliloti species induced a higher nodulation rate than S. medicae species (P = 0.01).
(ii) S. medicae mixture versus S. meliloti mixture effects.
When plant biomass means between S. medicae and S. meliloti mixtures were compared, a slight bacterial species effect was detected (P = 0.023) (Table 5, two intraspecific mixtures), with a higher biomass for plants inoculated with the mixed inoculant of S. medicae species.
(iii) Intraspecific mixtures versus single inoculants.
Within each species, dry aerial biomasses obtained with S. meliloti and S. medicae mixtures were either equivalent to or significantly higher than the mean of each single-strain inoculation (Fig. 2 and Table 5, S. medicae and S. meliloti). The S. medicae mixture and the md2 genotype induced similar plant biomasses, in contrast to the other three bacterial genotypes (Table 6). Furthermore, post hoc tests showed that the S. meliloti mixture induced greater aerial biomass than five genotypes (ml1, ml2, ml4, ml5, and ml7), whereas no significant difference was detected between the mixture and ml3 or ml6 for DZA315-16 and F83-005 and ml3 or ml8 for JemalongA17 (Table 6).
TABLE 6.
Comparisons of plant aerial biomass inoculated with each single bacterial genotype and S. medicae or S. meliloti intraspecific mixtures
| Inoculant | Bacterial genotype | Biomass comparison for plant linea:
|
|||
|---|---|---|---|---|---|
| DZA315-16 | PRT180-21 | F83-005 | JemalongA17 | ||
| S. medicae intraspecific mixture | md1 | <0.001 | <0.001 | <0.001 | <0.001 |
| md2 | 0.9249 | 0.4803 | 0.2018 | 0.6550 | |
| md3 | <0.001 | <0.001 | <0.001 | <0.001 | |
| md4 | ND | 0.0054 | <0.001 | <0.001 | |
| S. meliloti intraspecific mixture | ml1, ml2, ml4, ml5, ml7 | <0.05 | <0.05 | <0.05 | <0.05 |
| ml3 | 0.9607 | <0.001 | 0.0621 | 0.1799 | |
| ml6 | 0.2547 | <0.001 | 0.2509 | 0.0056 | |
| ml8 | <0.001 | <0.001 | <0.001 | 0.5200 | |
Based on Fisher post hoc tests (P values are shown). The numbers in boldface indicate that the genotypes were not significantly different from the bacterial mixtures considered. ND, not determined.
DISCUSSION
As previously found in several studies by studying the genetic diversity of rhizobia at five loci (11, 40), S. medicae contained less diversity than S. meliloti. Within each species, divergence among strains was detected on all loci in S. meliloti, whereas nucleotide variability was found only on nodulation loci for S. medicae. IGSNOD is characterized in both S. meliloti and S. medicae by negative Tajima's D values, which depart significantly from a neutral expectation for S. medicae, indicating that symbiotic nod locus genetic diversity is apparently actively maintained for both bacterial species by purifying selection. Since Tajima's D value could not be computed for typA and virB11 loci in S. medicae due to the lack of nucleotidic diversity, the negative D value for the IGSNOD locus might result from a demographic expansion. When the demographic exponential growth rates (G) of the two species are estimated with Lamarc software, S. medicae has a much higher value (Gestimated = 6,200) than S. meliloti (Gestimated = 11), implying that S. medicae may have had a much more rapid expansion rate than S. meliloti. Such demographic expansion would also fit with the much higher proportion in the soil observed for S. medicae than for S. meliloti.
Nucleotidic polymorphism was observed for the two homologous genes implicated in translocation and conjugation, virB11 and avhB11, between the two species, but also within S. meliloti. Our results support the hypothesis that the common ancestor of the two Sinorhizobium species harbored the two copies virB11 and avhB11. As several authors (2, 5) have suggested that S. medicae might have emerged from a population of S. meliloti, the emerging species S. medicae would have kept one ortholog, whereas S. meliloti harbored either the two copies or only one. The role of each gene in S. meliloti and between the two species merits more study.
In the trapping, we observed a high predominance of S. medicae strains compared to S. meliloti. In previous literature (2), no marked symbiotic preference for S. medicae was described for any M. truncatula line. Thus, the proportions observed in our study certainly reflect a major disequilibrium between the two bacterial species in the soil.
Medicago species display a wide variability of specificity (4). Such variations imply that Medicago species might play a role in bacterial selection in the soil, as demonstrated by Jebara et al. (19). In our study, the proportions of the two bacterial species (S. meliloti and S. medicae) were slightly different among the four plant lines at both interspecific and intraspecific levels, suggesting that the plant lines might select their symbiotic partners differently. M. truncatula plant lines thus display differences in their specificities against bacterial populations, although the presence or absence of rare strains might influence this differentiation. Consequently, M. truncatula genetic diversity in a natural ecosystem might, at least in part, have an influence on rhizobial genetic diversity in the soil, because viable bacteria could be released into the plant rhizosphere (25).
Both intraspecies competition results and soil-trapping results showed one genotype highly predominant for S. medicae (but not the same genotype) and a large genetic diversity for S. meliloti. Thus, a strong competition might exist among the different S. medicae genotypes for nodulation, leading to one predominant genotype being recovered, whereas this interstrain competition is apparently much weaker in S. meliloti, for which several genotypes were recovered in similar frequencies. We have no clue to explain such proportion differences in the competition outputs between the two species, even if it is not an intrinsic specificity of each species, as Bailly et al. (3) did not observe this unequal distribution in similar trapping experiments. Moreover, in S. medicae, the most competitive strain (md4) in the competition experiment was not the genotype found to be dominant in soil trapping (md1). We suspect that the md1 genotype is quantitatively dominant in the soil used but is not the most competitive for plant nodulation under the tube conditions tested, because competition among strains seems to be dependent on the experimental culture conditions, as previously demonstrated in the Bradyrhizobium japonicum-soybean model (15).
At an interspecies level, the competition experiment resulted in a higher proportion of S. meliloti, whereas trapping from soil resulted in more than 90% S. medicae isolated. The initial unbalanced proportions obtained in soil trapping most certainly resulted from a “population effect” rather than a methodological bias or a much higher competitiveness of S. medicae in soil. At least in this local population, we may consider S. meliloti the best M. truncatula symbiont for nodulation. To generalize this hypothesis, other competition analyses should be done between the two bacterial species trapped from other natural populations.
In Sinorhizobium spp., several authors have shown variation in the ability to fix nitrogen among strains originating from different geographical areas for S. meliloti (22, 27) or for S. medicae (17). In this study, we demonstrated high polymorphism in terms of nitrogen fixation effectiveness and nodulation ability within a single soil, at both the interspecies and the intraspecies levels, even though weak genotypic and nucleotidic divergence were observed among strains.
Within each bacterial species, we detected a significant bacterial genotype-plant line interaction on the aerial biomass and nodule number data. Previous studies showed that the fixation effectiveness of three S. meliloti strains (27) and two S. medicae strains (17) was dependent on the M. truncatula lines. Our study suggests that such interaction is observed not only among strains sampled from various geographical locations, but also within a single site, and that this can lead to dramatically divergent phenotypes (Fig. 2a, strain STM 5472). Nitrogen fixation variation among plant lines had already been described, and ecotype-strain specificity is, at least partially, controlled by a single recessive allele (Mtsym6) for the plant partner (39), whereas succinoglycan oligosaccharide structure is involved, at least partially, in the bacterial partner specificity (34).
The bacterial mixture containing the four S. medicae genotypes gave the same plant aerial biomass as the genotype md2, which is the most efficient of the four genotypes. The effectiveness reached by the mixture could thus be due to this single strain (md2), even if it was barely found in nodules. However, genotype occurrence could be underestimated, as coinfected nodules, which may represent around 15% (13), were not detected. In S. meliloti, depending on the plant line, either several genotypes displayed the same effectiveness as the mixture or the effectiveness of the mixture was found to be intermediate among all the strains. By comparing the effectiveness of each genotype separately, S. meliloti appears to be a better nitrogen-fixing species than S. medicae. Conversely, the bacterial mixture encompassing all S. medicae genotypes induces a better aerial plant biomass than the mixture including all S. meliloti strains. These opposite results might be explained by synergistic effects between strains in mixtures, as suggested by Bromfield (6), leading to a fixation advantage for S. medicae populations. This beneficial effect is not always the rule, as Heath and Tiffin (17) observed an antagonistic effect with a mixture of strains. Discrimination between S. meliloti and S. medicae as the best nitrogen-fixing symbiont species is not trivial, considering that in soil, populations of bacteria, rather than single strains, interact with the plant.
The influence of the plant cover on the structure evolution of soil bacterial populations appears extremely complex. Fine symbiotic differences exist within M. truncatula lines and can be expressed even when they are put in contact with a single soil. We showed that the partner choice in the M. truncatula-Sinorhizobium association is controlled by the plant lines and by the bacterial competition for nodulation and not strictly by the fixation efficiencies of the bacterial genotypes. However, it is relevant to keep in mind that bacterial adaptation to the environment is essential for symbiosis.
Acknowledgments
This work was supported by a program funded by INRA Santé des Plantes et Environnement (Rhizosphere Ecology of Annual Medics program). C. Rangin was supported by a Ph.D. fellowship from the French Ministry of Education and Research and by a teaching fellowship from the University Montpellier II.
We thank Lucette Mauré and Mélodie Bousquet, who provided technical help for strain isolation. We are very grateful to Christophe Mougel and Pierre Offre, who collected nodules from the soil-trapping experiment.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Arnaud-Haond, S., and K. Belkhir. 2007. Genclone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol. Ecol. Notes 7:15-17. [Google Scholar]
- 2.Bailly, X., I. Olivieri, B. Brunel, J.-C. Cleyet-Marel, and G. Bena. 2007. Horizontal gene transfer and homologous recombination drive the evolution of the nitrogen-fixing symbionts of Medicago species. J. Bacteriol. 189:5223-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailly, X., I. Olivieri, S. De Mita, J.-C. Cleyet-Marel, and G. Bena. 2006. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Mol. Ecol. 15:2719-2734. [DOI] [PubMed] [Google Scholar]
- 4.Bena, G., A. Lyet, T. Huguet, and I. Olivieri. 2005. Medicago-Sinorhizobium symbiotic specificity evolution and the geographic expansion of Medicago. J. Evol. Biol. 18:1547-1558. [DOI] [PubMed] [Google Scholar]
- 5.Biondi, E. G., E. Pilli, E. Giuntini, M. L. Roumiantseva, E. E. Andronov, O. P. Onichtchouk, O. N. Kurchak, B. V. Simarov, N. I. Dzyubenko, A. Mengoni, and M. Bazzicalupo. 2003. Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol. Lett. 220:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Bromfield, E. S. P. 1984. Variation in preference for Rhizobium meliloti within and between Medicago sativa cultivars grown in soil. Appl. Environ. Microbiol. 48:1231-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broughton, W. J., N. Heycke, U. Priefer, G.-M. Schneider, and J. Stanley. 1987. Ecological genetics of Rhizobium meliloti: diversity and competitive dominance. FEMS Microbiol. Lett. 40:245-249. [Google Scholar]
- 8.Cook, D. R. 1999. Medicago truncatula—a model in the making. Curr. Opin. Plant Biol. 2:301-304. [DOI] [PubMed] [Google Scholar]
- 9.De Lajudie, P., A. Willems, B. Pot, D. Dewettinck, G. Maestrojuan, M. Neyra, M. D. Collins, B. Dreyfus, K. Kersters, and M. Gillis. 1994. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int. J. Syst. Bacteriol. 44:715-733. [Google Scholar]
- 10.Depret, G., and G. Laguerre. 2008. Plant phenology and genetic variability in root and nodule development strongly influence genetic structuring of Rhizobium leguminosarum biovar viciae populations nodulating pea. New Phytol. 179:224-235. [DOI] [PubMed] [Google Scholar]
- 11.Eardly, B. D., L. A. Materon, N. H. Smith, D. A. Johnson, M. D. Rumbaugh, and R. K. Selander. 1990. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl. Environ. Microbiol. 56:187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 13.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garau, G., W. G. Reeve, L. Brau, P. Deiana, R. J. Yates, D. James, R. Tiwari, G. W. O'Hara, and J. G. Howieson. 2005. The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 276:263-277. [Google Scholar]
- 15.George, T., B. B. Bohlool, and P. W. Singleton. 1987. Bradyrhizobium japonicum-environment interactions: nodulation and interstrain competition in soils along an elevational transect. Appl. Environ. Microbiol. 53:1113-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geurts, R., E. Fedorova, and T. Bisseling. 2005. Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8:346-352. [DOI] [PubMed] [Google Scholar]
- 17.Heath, K. D., and P. Tiffin. 2007. Context dependence in the coevolution of plant and rhizobial mutualists. Proc. R. Soc. Lond. Ser. B 274:1905-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huss-Danell, K. 1980. Nitrogen fixation and biomass production in clones of alnus incana. New Phytol. 85:503-511. [Google Scholar]
- 19.Jebara, M., R. Mhamdi, M. E. Aouani, R. Ghrir, and M. Mars. 2001. Genetic diversity of Sinorhizobium populations recovered from different Medicago varieties cultivated in Tunisian soils. Can. J. Microbiol. 47:139-147. [DOI] [PubMed] [Google Scholar]
- 20.Kiss, E., T. Huguet, V. Poinsot, and J. Batut. 2004. The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobium meliloti 1021 with certain Medicago truncatula lines. Mol. Plant-Microbe Interact. 17:235-244. [DOI] [PubMed] [Google Scholar]
- 21.Meade, J., P. Higgins, and F. O'Gara. 1985. Studies on the inoculation and competitiveness of a Rhizobium leguminosarum strain in soils containing indigenous rhizobia. Appl. Environ. Microbiol. 49:899-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, R. W., and J. C. Sirois. 1982. Relative efficacy of different alfalfa cultivar-Rhizobium meliloti strain combinations for symbiotic nitrogen fixation. Appl. Environ. Microbiol. 43:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mnasri, B., M. Mrabet, G. Laguerre, M. E. Aouani, and R. Mhamdi. 2007. Salt-tolerant rhizobia isolated from a Tunisian oasis that are highly effective for symbiotic N2-fixation with Phaseolus vulgaris constitute a novel biovar (bv. mediterranense) of Sinorhizobium meliloti. Arch. Microbiol. 187:79-85. [DOI] [PubMed] [Google Scholar]
- 24.Mytton, L. R., M. H. El-Sherbeeny, and D. A. Lawes. 1977. Symbiotic variability in Vicia faba. 3. Genetic effects of host plant, rhizobium strain and of host × strain interaction. Euphytica 26:785-791. [Google Scholar]
- 25.Paau, A. S., C. B. Bloch, and W. J. Brill. 1980. Developmental fate of Rhizobium meliloti bacteroids in alfalfa nodules. J. Bacteriol. 143:1480-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paffetti, D., F. Daguin, S. Fancelli, S. Gnocchi, F. Lippi, C. Scotti, and M. Bazzicalupo. 1998. Influence of plant genotype on the selection of nodulating Sinorhizobium meliloti strains by Medicago sativa. Antonie van Leeuwenhoek 73:3-8. [DOI] [PubMed] [Google Scholar]
- 27.Parra-Colmenares, A., and M. L. Kahn. 2005. Determination of nitrogen fixation effectiveness in selected Medicago truncatula isolates by measuring nitrogen isotope incorporation into pheophytin. Plant Soil 270:159-168. [Google Scholar]
- 28.Raymond, M., and F. Rousset. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86:248-249. [Google Scholar]
- 29.Rome, S., M. P. Fernandez, B. Brunel, P. Normand, and J. C. Cleyet-Marel. 1996. Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int. J. Syst. Bacteriol. 46:972-980. [DOI] [PubMed] [Google Scholar]
- 30.Ronfort, J., T. Bataillon, S. Santoni, M. Delalande, J. David, and J.-M. Prosperi. 2006. Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula. BMC Plant Biol. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roumiantseva, M. L., E. E. Andronov, L. A. Sharypova, T. Dammann-Kalinowski, M. Keller, J. P. Young, and B. V. Simarov. 2002. Diversity of Sinorhizobium meliloti from the Central Asian alfalfa gene center. Appl. Environ. Microbiol. 68:4694-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shishido, M., and I. L. Pepper. 1990. Identification of dominant indigenous Rhizobium meliloti by plasmid profiles and intrinsic antibiotic resistance. Soil Biol. Biochem. 22:11-16. [Google Scholar]
- 33.Silva, C., F. L. Kan, and E. Martínez-Romero. 2007. Population genetic structure of Sinorhizobium meliloti and S. medicae isolated from nodules of Medicago spp. in Mexico. FEMS Microbiol. Ecol. 60:477-489. [DOI] [PubMed] [Google Scholar]
- 34.Simsek, S., T. Ojanen-Reuhs, S. B. Stephens, and B. L. Reuhs. 2007. Strain-ecotype specificity in Sinorhizobium meliloti-Medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J. Bacteriol. 189:7733-7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyman, C. P., and B. W. Strijdom. 1980. Symbiotic characteristics of lines and cultivars of Medicago truncatula inoculated with strains of Rhizobium meliloti. Phytophylatica 12:173-176. [Google Scholar]
- 36.Sunnucks, P., A. C. Wilson, L. B. Beheregaray, K. Zenger, J. French, and A. C. Taylor. 2000. SSCP is not so difficult: the application and utility of single-stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol. Ecol. 9:1699-1710. [DOI] [PubMed] [Google Scholar]
- 37.Thurman, N. P., and E. S. P. Bromfield. 1988. Effect of variation within and between Medicago and Melilotus species on the composition and dynamics of indigenous populations of Rhizobium meliloti. Soil Biol. Biochem. 20:31-38. [Google Scholar]
- 38.Tian, C., E. Wang, T. Han, X. Sui, and W. Chen. 2007. Genetic diversity of rhizobia associated with Vicia faba in three ecological regions of China. Arch. Microbiol. 188:273-282. [DOI] [PubMed] [Google Scholar]
- 39.Tirichine, L., F. de Billy, and T. Huguet. 2000. Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiol. 123:845-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Berkum, P., Y. Badri, P. Elia, M. E. Aouani, and B. D. Eardly. 2007. Chromosomal and symbiotic relationships of rhizobia nodulating Medicago truncatula and M. laciniata. Appl. Environ. Microbiol. 73:7597-7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications Ltd., Oxford, United Kingdom.
- 42.Yates, R. J., J. G. Howieson, D. Real, W. G. Reeve, A. Vivas-Marfisi, and G. W. O'Hara. 2005. Evidence of selection for effective nodulation in the Trifolium spp. symbiosis with Rhizobium leguminosarum biovar trifolii. Aust. J. Exp. Agric. 45:189-198. [Google Scholar]
- 43.Zribi, K., R. Mhamdi, T. Huguet, and M. E. Aouani. 2004. Distribution and genetic diversity of rhizobia nodulating natural populations of Medicago truncatula in Tunisian soils. Soil Biol. Biochem. 36:903-908. [Google Scholar]