Abstract
Multilocus sequence typing and fimA genotyping were performed on Porphyromonas gingivalis isolates from 15 subjects with “refractory” periodontitis. Several sequence types were detected for most individual pockets. The variation indicated recombination at the recA and pepO genes. The prevalence of fimA genotypes II and IV confirmed their association with periodontitis.
Multilocus sequence typing (MLST) is currently the best tool for epidemiological studies of most bacterial pathogens and for understanding population structure (5, 10, 31). An MLST scheme for the periodontal pathogen Porphyromonas gingivalis (8, 9, 23) has been developed, and sequence types (STs) from human periodontitis isolates are accessible in the MLST database (www.pubmlst.org/pgingivalis) (22).
P. gingivalis, a gram-negative black-pigmented anaerobic rod, residing in subgingival biofilms, is widely recognized as a contributor to the development of periodontitis together with other oral pathogens (17, 18). The species has also been reported to cause extraoral infections (27, 38, 39).
P. gingivalis' major fimbriae are important in adherence to host cells and other surfaces (17, 18, 24, 25) and are classified into six types (I, Ib, II, III, IV, and V) based on sequence variation of the fimA gene (2, 6, 14, 34, 35). Several PCR studies applied with clinical material and genotyping of cultured periodontitis isolates indicate that fimA genotypes II, Ib, and IV are more frequently associated with disease than other genotypes (2, 4, 8, 33-35).
The aims of this study were to detect by culturing the prevalence of P. gingivalis in a single diseased site in a number of patients with “refractory” periodontitis and to characterize the isolates of the species by MLST and fimA genotyping (3, 15, 40, 41).
One periodontal pocket with clinical signs of inflammation (pocket depths of 6 mm or more and bleeding on probing [score 1]) (26) was selected for microbiological sampling for each of 30 subjects (19, 28). According to self-reporting and dental records, antibiotics had not been used for 6 months. Sterile paper points (37) were inserted in the site for 15 s before being put into a prereduced anaerobically sterilized dental transport medium (Anaerobe Systems, Morgan Hill, CA). One specialist in periodontology (M.E.), who treated the patients, performed the diagnostics. The project was approved by the regional committee for medical research ethics (project no. 2.2005.2667).
From each sample, 100 μl vortexed transport medium was plated on a Columbia agar plate under continuous N2 flow and incubated anaerobically for 5 to 7 days (90% N2, 5% H2, 5% CO2) at 37°C (WS9000; Anoxomat, Mart, The Netherlands).
For the primary plates of 15 patients, black-pigmented bacteria constituted 75 to 100% of the growth. Using an inoculation loop (1 μl), mainly black-pigmented colonies were spread on a fresh Columbia agar plate. This was repeated 5 to 10 times to randomly pick colonies from different parts of the primary plates. The new plates were then used to obtain single colonies. Purity was inspected under a stereo microscope (Stemi SV6; 8× to 50×; Zeiss), and selection of single colonies was done randomly. When pure, they were replated and incubated for another 10 to 14 days. A total of 93 isolates from 2 to 10 colonies per patient site were obtained and further investigated.
DNA was extracted from bacterial cultures, using a Magnatrix 1200 biomagnetic workstation (Magnetic Biosolutions, Stockholm, Sweden) and the MagAttract DNA Mini M48 kit (Qiagen, California), according to the manufacturers' instructions.
Sequencing of the 16S rRNA gene was performed to ascertain species identification. All 93 isolates further studied showed 99 to 100% sequence identity to the 16S rRNA gene of P. gingivalis from GenBank. fimA genotyping and MLST were performed as described previously (8, 9, 23). Construction of the unweighted pair group method with arithmetic averages (UPGMA) dendrogram (21) and the splits decomposition analysis were computed (20). The eBurst analysis was done using the eBurst, version 3, software program (http://eburst.mlst.net) (11, 42).
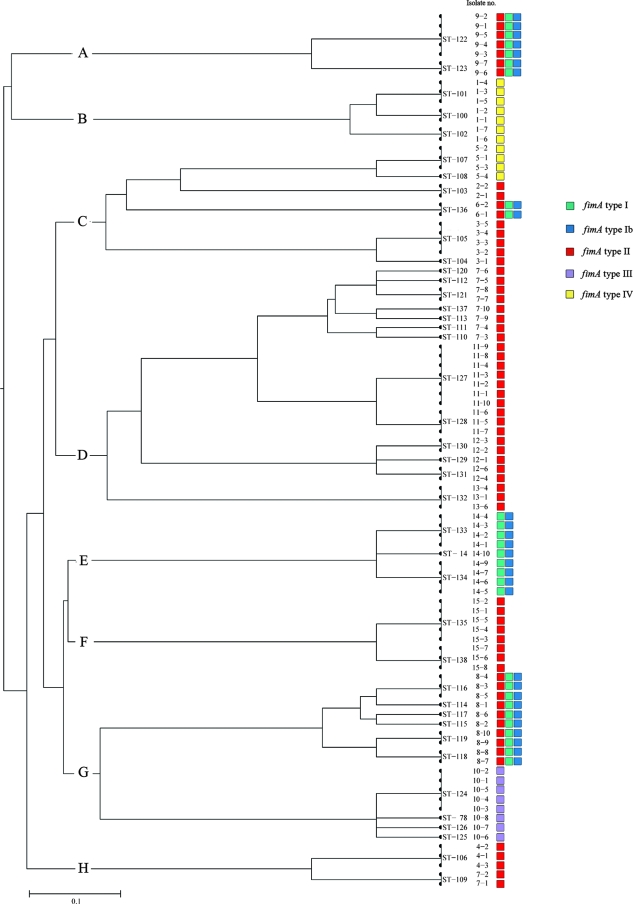
The results of the fimA genotyping are shown in Fig. 1. For each subject, all isolates showed identical fimA genotypes. Forty-six isolates (49.5%) from eight subjects were fimA genotype II, eleven isolates (11.8%) from two subjects were genotype IV, and eight isolates (8.6%) from one subject were genotype III.
FIG. 1.
Genetic relationships among 93 P. gingivalis isolates. A distance matrix using UPGMA was calculated on the basis of differences in allelic profiles. Clusters at a genetic distance of 0.40 were designated A to H. The STs, strain designations, and fimA genotypes of the isolates are indicated.
Genotypes I, Ib, and V alone were not observed (Fig. 1). The remaining 28 (30.1%) isolates showed positive PCR with more than one primer set and were either I, Ib, and II (three patients) or I and Ib (one patient). The 93 isolates were assigned to 41 STs, of which 39 were new. ST14 and ST78 had been identified previously for German patients (23). From 1 to 8 STs were found among the colonies from a single pocket in these 15 subjects with “refractory” periodontitis.
The UPGMA dendrogram (Fig. 1) showed evolutionary relationships among the STs. Eight clusters were observed (A to H) at a linkage distance of 0.40. Clusters A, B, E, and F contained the isolates collected from single subjects, 1, 9, 14, and 15. Clusters C, D, G, and H harbored isolates from several subjects (Fig. 1).
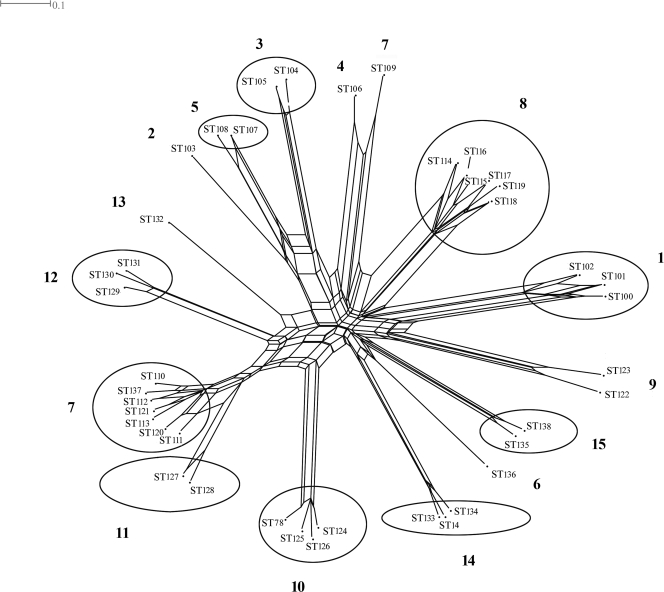
The results of the eBURST analysis identified 10 clonal complexes defined by single locus variants to single sites superimposed on the obtained SplitsTree graph (Fig. 2).
FIG. 2.
Degree of recombination between 93 isolates of P. gingivalis for the seven gene fragments in the MLST, illustrated by the SplitsTree software program, version 4.6, downloaded from http://www.splitstree.org/. Clonal complexes (single locus variants) are encircled. Subject numbers are in bold.
Only closely related STs were observed in individual pockets, and except for one case (subject 7), variation within a site was limited to the pepO and recA genes.
When more than three isolates within a site were analyzed, several alleles were distinguished, which differed in two, three, or four single nonadjacent nucleotides (shown by ClustalW alignments). For subject 7, the allelic profile of ST109 presented different alleles for most genes compared to those of the other isolates.
The results of the specific fimA genotyping were superimposed on the dendrogram (Fig. 1) and showed no variation for each subject, even when the isolates belonged to several STs.
This study of the genetic diversity among 93 P. gingivalis isolates from single “refractory” periodontitis sites in 15 Norwegian patients supported earlier findings of the large heterogeneity of the species (1, 8, 9, 12, 30, 32). We reported previously the occurrence of several STs within three Indonesian patients (8, 9), but we demonstrated here that single sites can be colonized by multiple STs. Up to 8 STs were identified in 1 site with a maximum of 10 isolates analyzed. The findings are noteworthy considering the general acceptance that individuals with periodontitis harbor only one clone (29, 40).
The fact that P. gingivalis can be detected in diseased periodontal sites and in healthy gingival sulci (7, 36, 40) suggests that it is a species of variable pathogenic potential. Therefore, diseased sites have been hypothesized to be colonized by more-virulent clones.
Since all sites investigated showed signs of severe inflammation, analysis of isolates of P. gingivalis at the sites by using MLST and fimA genotyping could provide a characterization of the most virulent clones (40). It was noteworthy that two STs (ST14 and ST78) identified in this study were previously recorded in the MLST isolate database (www.pubmlst.org/pgingivalis), representing isolates sampled from aggressive periodontitis cases in Germany. These isolates may correspond to more-virulent clones of the species.
With the exception of subject 7, the variation in each site was related only to the pepO and recA genes. The multiple alleles of the pepO and recA genes at individual sites differed in nonadjacent nucleotides and could be signs of recombination. This is in accordance with other studies that have concluded that recombination occurs to a larger extent among bacterial species than previously thought (13, 16) and in segments of chromosomal DNA larger than the fragments analyzed here. Variation in the different alleles occurred at a limited number of nucleotides, suggesting recombination occurring within the periodontal site.
It is unlikely that multiple point mutations would arise in the same positions in the different alleles involved.
The function of the gene upstream and next to the pepO gene (PG0145; coding for the purine/pyrimidine phosphoribosyl transferase enzyme, related to DNA transformation) and the function of the recA gene itself (PG0789; DNA metabolism; DNA replication, recombination, and repair) (http://www.oralgen.lanl.gov/) may explain why these genes are more subject to recombination than the other MLST genes. In a recent study, Tribble et al. (43) demonstrated the ability of different strains of P. gingivalis to transfer chromosomal DNA to each other by conjugation, which is postulated to be an underlying mechanism for allele swapping and genetic variation in the species. Indications of frequent recombination have also been provided by Frandsen et al. (12), who found a random distribution of two virulence-associated mobile genetic elements.
The fimA genotyping of the isolates investigated demonstrated a high prevalence of fimA genotypes II and IV and supports earlier findings of the association between the genotypes and disease (2, 8, 33).
Acknowledgments
We are indebted to the Faculty of Dentistry, University of Oslo, Oslo, Norway, for financial support.
We thank Emnet Abesha-Belay, Merete Sandvik, Isabelle Messel, Anne-Marie Klem, and Jan Oksnes for technical assistance.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Ali, R. W., L. Martin, A. D. Haffajee, and S. S. Socransky. 1997. Detection of identical ribotypes of Porphyromonas gingivalis in patients residing in the United States, Sudan, Romania and Norway. Oral Microbiol. Immunol. 12:106-111. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 37:1426-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage, G. C. 1999. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 4:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Beikler, T., U. Peters, S. Prajaneh, K. Prior, B. Ehmke, and T. F. Fleming. 2003. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur. J. Oral Sci. 111:390-394. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, J. E., and E. J. Feil. 2004. Multilocus sequence typing—what is resolved? Trends Microbiol. 12:373-377. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson, D. P., M. A. Kubiniec, F. Yoshimura, and R. J. Genco. 1988. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J. Bacteriol. 170:1658-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316-323. [DOI] [PubMed] [Google Scholar]
- 8.Enersen, M., I. Olsen, Ø. Kvalheim, and D. A. Caugant. 2008. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from periodontitis. J. Clin. Microbiol. 46:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enersen, M., I. Olsen, A. J. van Winkelhoff, and D. A. Caugant. 2006. Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J. Clin. Microbiol. 44:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frandsen, E. V., K. Poulsen, M. A. Curtis, and M. Kilian. 2001. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect. Immun. 69:4479-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, C., W. P. Hanage, and B. G. Spratt. 2007. Recombination and the nature of bacterial speciation. Science 315:476-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara, T., S. Morishima, I. Takahashi, and S. Hamada. 1993. Molecular cloning and sequencing of the fimbrillin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem. Biophys. Res. Commun. 197:241-247. [DOI] [PubMed] [Google Scholar]
- 15.Haffajee, A. D., N. G. Uzel, E. I. Arguello, G. Torresyap, D. M. Guerrero, and S. S. Socransky. 2004. Clinical and microbiological changes associated with the use of combined antimicrobial therapies to treat “refractory” peridontitis. J. Clin. Periodontol. 31:869-871. [DOI] [PubMed] [Google Scholar]
- 16.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. The impact of homologous recombination on the generation of diversity in bacteria. J. Theor. Biol. 239:210-219. [DOI] [PubMed] [Google Scholar]
- 17.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 18.Holt, S. C., and J. L. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72-122. [DOI] [PubMed] [Google Scholar]
- 19.Horz, H. P., and G. Conrads. 2007. Diagnosis and anti-infective therapy of periodontitis. Expert Rev. Anti Infect. Ther. 4:703-715. [DOI] [PubMed] [Google Scholar]
- 20.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 21.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 22.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koehler, A., H. Karch, T. Beikler, T. F. Flemmig, S. Suerbaum, and H. Schmidt. 2003. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology 149:2407-2415. [DOI] [PubMed] [Google Scholar]
- 24.Lamont, R. J., and H. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 26.Lang, N. P., A. Joss, and M. Tonetti. 1996. Monitoring disease during supportive periodontal treatment by bleeding on probing. Periodontol. 2000 12:44-48. [DOI] [PubMed] [Google Scholar]
- 27.Li, X., K. M. Kolltveit, L. Tronstad, and I. Olsen. 2000. Systemic diseases caused by oral infection. Clin. Microbiol. Rev. 13:547-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Listgarten, M., and P. M. Loomer. 2003. Microbial identification in the management of periodontal diseases. A systematic review. Ann. Periodontol. 8:182-192. [DOI] [PubMed] [Google Scholar]
- 29.Loos, B. G., A. J. Van Winkelhoff, R. G. Dunford, R. J. Genco, J. De Graaff, D. P. Dickinson, and D. W. Dyer. 1992. A statistical approach to the ecology of Porphyromonas gingivalis. J. Dent. Res. 71:353-358. [DOI] [PubMed] [Google Scholar]
- 30.Loos, B. G., D. W. Dyer, T. S. Whittam, and R. K. Selander. 1993. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect. Immun. 61:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menard, C., and C. Mouton. 1995. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect. Immun. 63:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Missailidis, C. G., J. E. Emeda, C. Ota-Tsuzuki, D. Anzai, and M. P. A. Mayer. 2004. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various forms of periodontal conditions. Oral Microbiol. Immunol. 19:224-229. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa, I., A. Amano, R. K. Kimura, T. Nakamura, S. Kawabata, and S. Hamada. 2000. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J. Clin. Microbiol. 38:1909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagawa, I., A. Amano, Y. Ohara-Nemoto, N. Endoh, I. Morisaki, S. Kimura, S. Kawabata, and S. Hamada. 2002. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J. Periodontal Res. 37:425-432. [DOI] [PubMed] [Google Scholar]
- 36.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Schlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 37.Renvert, S., M. Wikstrom, M. Helmersson, G. Dahlen, and N. Claffey. 1992. Comparative study of subgingival microbiological sampling techniques. J. Periodontol. 63:797-801. [DOI] [PubMed] [Google Scholar]
- 38.Scannapieco, F. A., R. B. Bush, and S. Paju. 2003. Association between periodontal disease and risk for nosocomial bacterial pneumonia and chronic obstructive pulmonary disease. A systematic review. Ann. Periodontol. 8:54-69. [DOI] [PubMed] [Google Scholar]
- 39.Scannapieco, F. A. 2005. Systemic effects of periodontal diseases. Dent. Clin. North Am. 49:533-550. [DOI] [PubMed] [Google Scholar]
- 40.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 41.Socransky, S. S., C. Smith, and A. D. Haffajee. 2002. Subgingival microbial profiles in refractory periodontal disease. J. Clin. Periodontol. 29:260-268. [DOI] [PubMed] [Google Scholar]
- 42.Spratt, B. G., W. P. Hanage, B. Li, D. Aanensen, and E. Feil. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 4:129-134. [DOI] [PubMed] [Google Scholar]
- 43.Tribble, G. A., G. J. Lamont, A. Progulske-Fox, and R. J. Lamont. 2007. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J. Bacteriol. 189:6382-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]