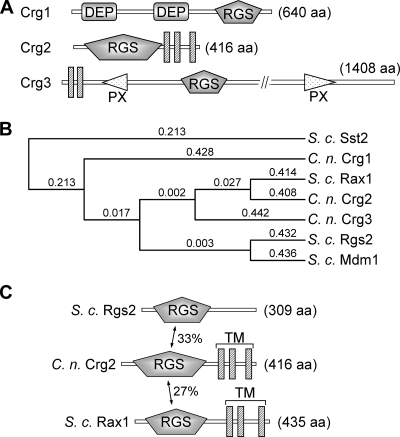
FIG. 1.
C. neoformans encodes three RGS protein homologs, Crg1, Crg2, and Crg3. (A) Schematic representations of three RGS proteins in C. neoformans. DEP, domains found in Dishevelled, Egl-10, and pleckstrin; PX, domains that bind to phosphoinositides; aa, amino acids. (B) The alignment of C. neoformans (C. n.) and S. cerevisiae (S. c.) RGS proteins indicates higher amino acid sequence similarity between Crg2 and Rax1. Protein sequences were aligned, and the phylogenic tree was drawn using MacVector software (MacVector, Inc.) with the following parameters: open gap penalty of 10.0, extend gap penalty of 0.2, delay divergence of 30%, gap distance of 4, and similarity matrix blosum. (C) Comparison of Crg2 with S. cerevisiae (S. c.) Rgs2 and Rax1 proteins. Crg2 contains an N-terminal RGS domain and three C-terminal transmembrane domains (TM). The GenBank accession numbers for Crg1, Crg2, Crg3, Sst2, Rgs2, Rax1, and Mdm1 are AAR06255, XP_570417, XP_569481, NP_013557, NP_014750.1, NP_014945.1, and NP_013603.1, respectively.