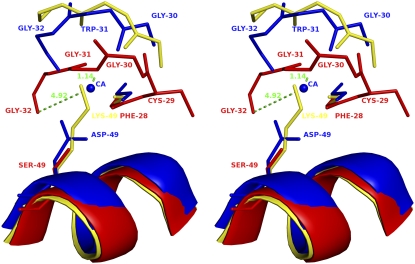
FIGURE 3.
Stereo diagram of superposition of Ca2+-binding loop of PLA2 enzymes. ScPLA2 (Protein Data Bank code, 1OZ6; blue), ecarpholin S (red), and myotoxin II (Protein Data Bank code, 1CLP; yellow) were superimposed. Conformational differences of three different Ca2+-binding loops are shown. Three residues Gly30-Trp31-Gly32 of scPLA2 bend to form a coordination bond with Ca2+, whereas three residues Gly30-Gly31-Gly32 of ecarpholin S extend linearly, causing Gly32 carbonyl O to move away from possible Ca2+ area (∼5 Å). The whole loop moves closer to Ser49 residue, and Nα of Gly31 nearly occupies putative Ca2+-binding position (1.1 Å away from Gly31 N), whereas in myotoxin II, Nɛ of Lys49 residue occupies Ca2+ position. The side-chain of Ser49 residue of ecapholin S is too short to extend into Ca2+-binding site (Oγ of Ser49 is 3.8 Å away from putative Ca2+-binding site). Image was prepared using the program Pymol (59).