Figure 7.
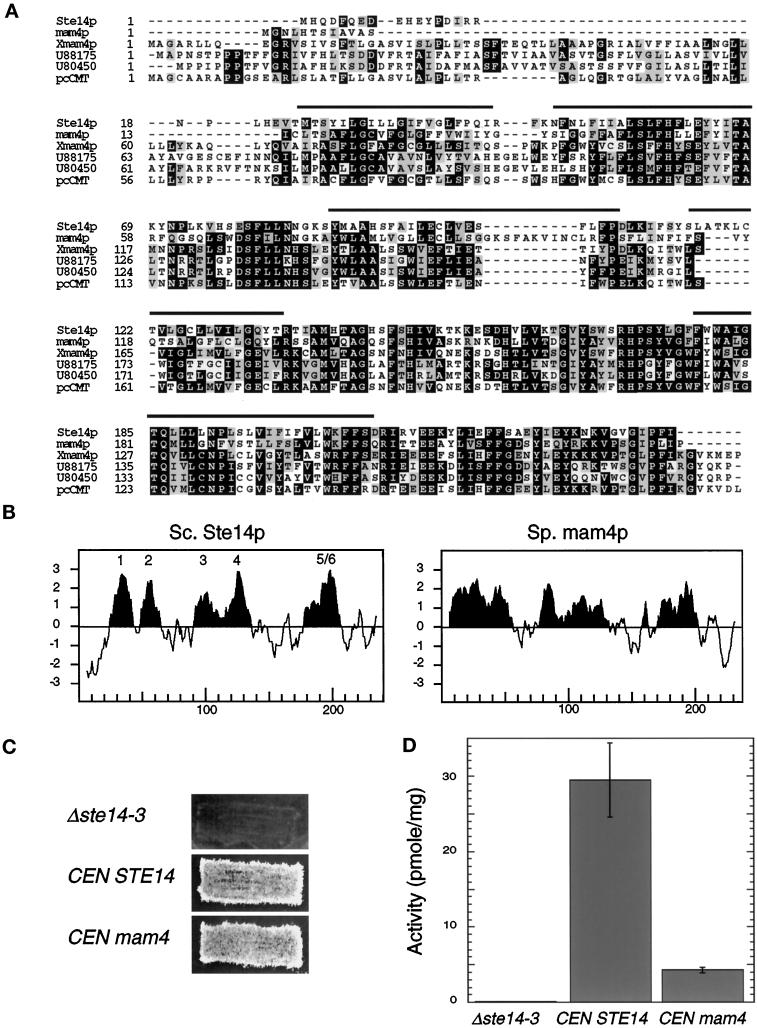
A S. pombe homologue complements a Δste14 defect. (A) Alignments of S. cerevisiae Ste14p, S. pombe mam4p, X. laevis Xmam4p, two C. elegans open reading frames (accession numbers U88175 and U80450), and human pcCMT are shown. Protein sequences were aligned using ClustalW software (Thompson et al., 1994). Black boxes denote amino acid identity, and gray boxes denote amino acid similarity as determined with the Boxshade server. The bars above the sequence denote the putative membrane spans of Ste14p. (B) Comparison of the hydropathy plots of Ste14p and mam4p is shown. Hydropathy plots were generated according to the algorithm of Kyte and Doolittle (1982) with a window of 11 amino acids. Hydrophobic regions are shown in black, and the potential membrane spans of Ste14p are indicated. The hydropathy profile of S. cerevisiae Ste14p and of S. pombe mam4p was calculated using the data of Sapperstein et al. (1994) and of Imai et al. (1997), respectively. (C) The coding sequence of S. pombe mam4 was expressed from the S. cerevisiae STE14 promoter in SM1188 (Δste14-3) and assayed for complementing activity by the plate mating assay (as in Figure 5C). Strains tested were SM2926 (Δste14-3), SM3185 (Δste14-3,CEN STE14), and SM3583 (Δste14-3,CEN mam4). (D) Methyltransferase activity of Δste14, Ste14p, and mam4p is shown. Each reaction performed in triplicate contained 5 μg of membrane proteins, 100 μM AFC, and 0.7 μM [3H]AdoMet and was incubated at 30°C for 30 min. The mean activity ± SD are shown.