Figure 6.
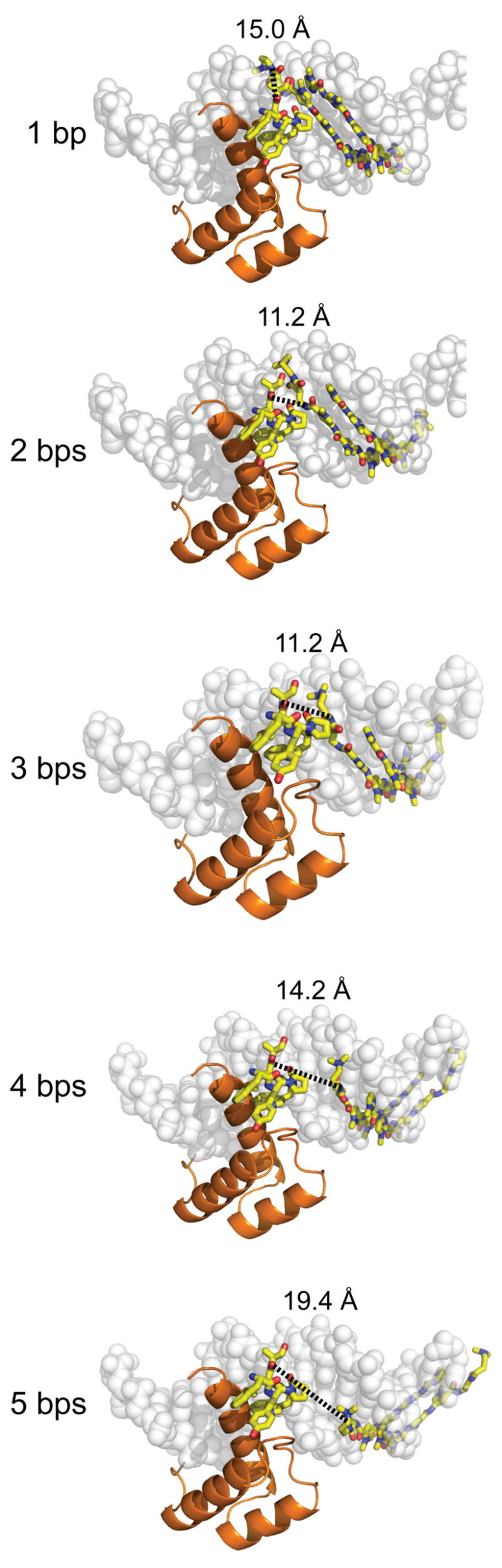
A series of estimated distance measurements are shown between the polyamide tail and the methionine carbonyl of the YPWM protein-binding domain which each dimerizer linker domain must traverse. The models of the polyamides binding to the DNA were generated by superimposing the DNA-binding site from a representative polyamide-DNA crystal structure (PDB code = 365D) over the Ubx/Exd/DNA crystal structure (PDB code 1B8I) and deleting the Ubx protein except for its YPWM protein-binding domain. In the proximal orientation, there exists a minimum distance of ~11 Å between the site spacings of 2–3 bps which increases non-linearly in each direction as the polyamide slides along the helical DNA minor groove.
