Fig. 1. Expression cloning of the R. leguminosarum lipid A oxidase in R. etli.
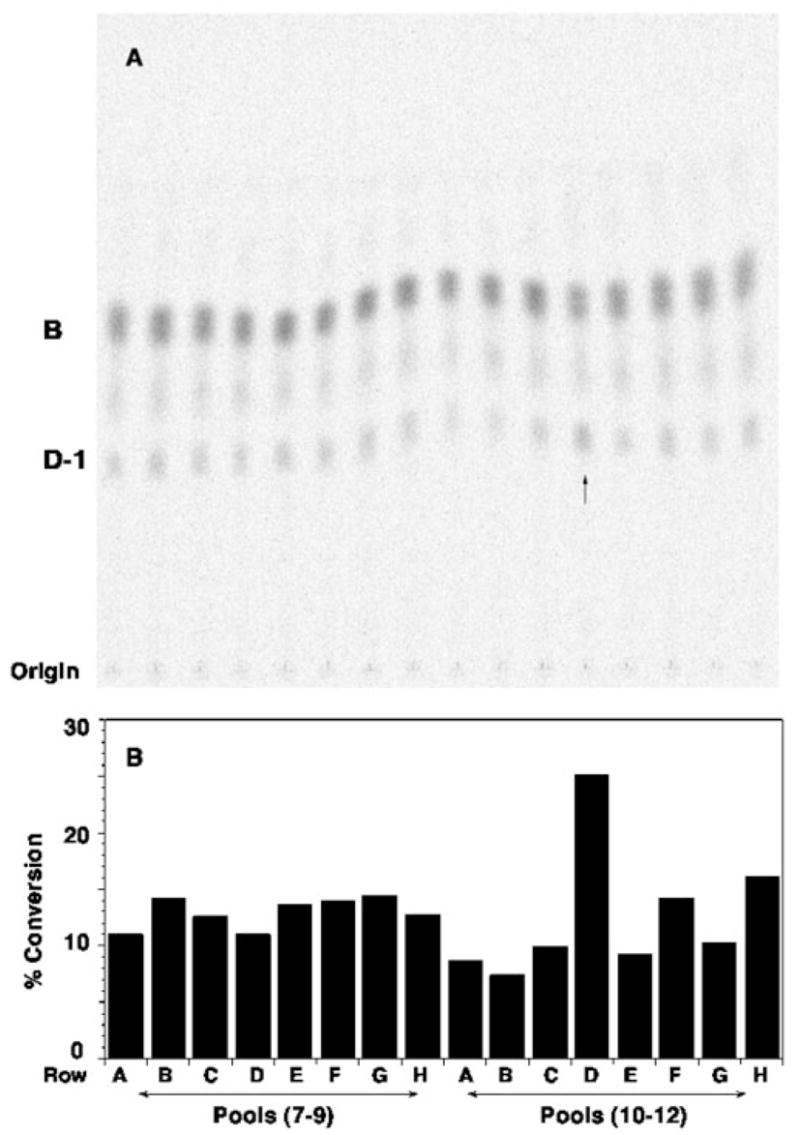
A, a genomic R. leguminosarum DNA library in cosmid pLAFR-1 (24, 32, 44) was transferred into R. etli CE3. Cells from ~1800 cosmid-containing colonies were grown up individually in 96-well microtiter plates. As described under “Experimental Procedures,” the lysates prepared by lysozyme treatment were pooled into groups of three and assayed for overexpression of the lipid A oxidase activity, as judged by the conversion of ~0.003 μM [14C]B (~600 cpm/reaction tube) to [14C]D-1 after 60 min. The arrow indicates a possible pool with elevated oxidase activity. B, the calculated percentage of conversion of [14C]B to [14C]D-1 is shown for each lane. The activity in the pool from row D (wells 10 –12 from plate 1S containing lysates 1S10D, 1S11D, and 1S12D) is approximately twice that of the other pools. Assays of the individual lysates (data not shown) revealed that only 1S11D contained high levels of oxidase activity. Among the ~1800 colonies tested, only three positive cosmids (p1S11D, p1E11D, and p1U12G) were identified in this manner. Based on restriction enzyme digests, p1S11D and p1E11D contained the same insert.
