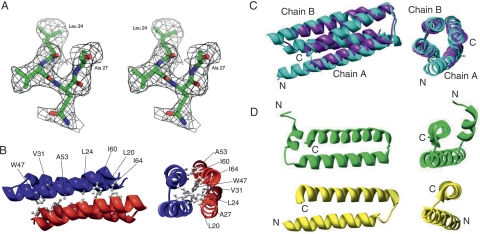
Figure 1.
Crystal structure and the simulated-annealing F o –F c omit map in the conserved region of AscE. (A) The map is contoured at a level of 2.0 σ. Residues Leu24 to Ala27 and all atoms within 3.0 Å of Leu24 to Ala27 were omitted prior to refinement. (B) Ribbon representation of the crystal structure of the dimeric AscE from residue Pro14 to Glu65 at two different angles. The hydrophobic residues (Leu20, Leu24, Ala27, Val31, Trp47, Ala53, Ile60, and Ile64) that form an interlocking network at the dimeric interface of the protein are shown in a ball-and-stick model. The figure was generated with the program Chimera (Pettersen et al. 2004). (C) Overlay of the crystal structures of AscE (purple) and YscE (chain A and B) (cyan) viewed at two different angles. The dimers of AscE and YscE overlay with an RMSD of 2.2 Å for 101 Cα atoms using DaliLite pairwise comparison of protein structure. (D) Ribbon representation of the structures of PscE (green) and YscE (yellow) as in the crystal structures of the complexes PscE-PscF55–85-PscG and YscEFG viewed at two different angles (Quinaud et al. 2007; Sun et al. 2008). Figure 1A was prepared using the program PyMOL (DeLano Scientific). Figure 1B–D was prepared using the program Chimera (Pettersen et al. 2004).