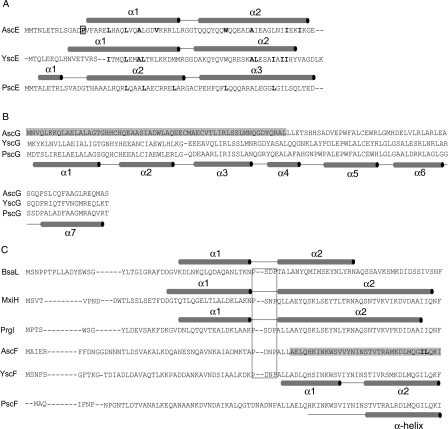
Figure 2.
Protein sequence alignment of A. hydrophila AH-1. Protein sequences of (A) AscE, (B) AscG, and (C) AscF were aligned with related proteins from Pseudomonas aeruginosa (PscE, PscG, and PscF), Yersinia pestis (YscE, YscG, and YscF), Shigella flexneri (MxiH), Salmonella typhimurium (PrgI), and Burkholderia pseudomallei (BsaL) using CLUSTLAW (Thompson et al. 1994). (A) The secondary structures as determined in the crystal structures of AscE, YscEFG, and PscE-PscF55-85-PscG are shown above their respective protein sequences. The hydrophobic residues in the dimeric interface are bold-faced and residue Pro14, which is conserved among AscE in Aeromonas species is bold-faced and boxed. (B) The proposed AscE binding region of AscG is shaded in gray. The secondary structure of PscG as determined in the complex formed between PscE, PscF55–85, and PscG is shown below the protein sequence of PscG. (C) Secondary structures as determined in the NMR structures of BsaL and PrgI and the crystal structure of MxiH are shown above their protein sequences. The conserved P-(S/D)-(D/N)-P motifs that form the interhelical turn are boxed. The proposed chaperone region of AscF is shaded in gray and the two hydrophobic residues (Ile83 and Leu84) of the C-terminal “ILQKI” sequence that are essential for chaperone binding are shown in bold-face type. The secondary structure of YscF as determined in the YscEFG complex is shown below the protein sequence of YscF. The secondary structure of PscF as determined in the complex formed between PscE, PscF55-85, and PscG is shown below the protein sequence of PscF.