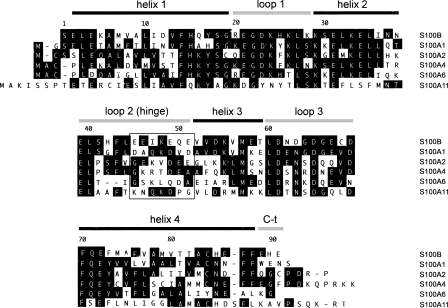
Figure 1.
Sequence comparison of S100 proteins. Alignment of the human S100 sequences studied. Sequences were aligned using Megalin (Laser Gene) software. Residues matching the consensus sequence are shaded. The secondary structure of calcium-bound S100B is indicated above the sequence. The net charge in the area delimited by a square in the loop 2 (hinge) region is negative in the proteins that bind to NRD peptide (S100B, S100A1, and S100A2) but zero or positive for those proteins that do not bind (or bind very weakly) to NRD peptide (S100A4, S100A6, and S100A11). This area contains negatively charged residues in S100B (E46, E49, E51), whose HSQC correlations were perturbed upon binding of NRD but not upon binding of TET-L348A (see Fig. 5B).