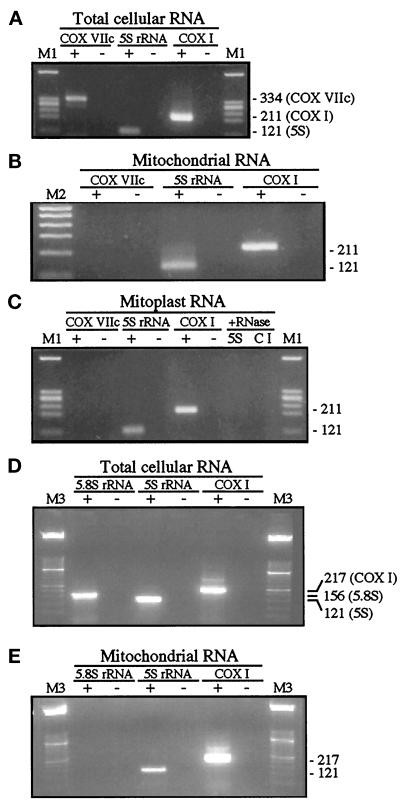
Figure 3.
RT-PCR analyses. (A) Analysis of 5S rRNA, COX I mRNA, and COX VIIc mRNA transcripts in total cellular RNA isolated from human 143B.TK− cells. RNA was subjected to RT-PCR in the presence (+) or absence (-) of reverse transcriptase activity. M1, markers of HaeIII-digested ΦX174 DNA. Expected sizes of RT-PCR products, in bp, are shown on the right. (B) Analysis described in A except that RNA from highly purified RNAse A–treated mitochondria was used as the template after DNase I treatment. M2, 100-bp ladder (Genosys Biotechnologies, The Woodlands, TX). (C) Analysis described in B except that RNA from highly purified RNase A–treated mitoplasts (obtained by the digitonin method) was used as the template. Mitoplasts were also lysed with 0.5% SDS in the presence of RNase A and subjected to RT-PCR to amplify 5S rRNA (5S) and COX I mRNA (CI). (D) Analysis of 5S rRNA, COX I mRNA, and 5.8S rRNA transcripts in total RNA isolated from rat liver. Methods and notation are described in A. M3, marker XIII (Boehringer Mannheim). (E) Analysis described in D except that RNA from highly purified RNAse A–treated rat liver mitochondria was used as the template.