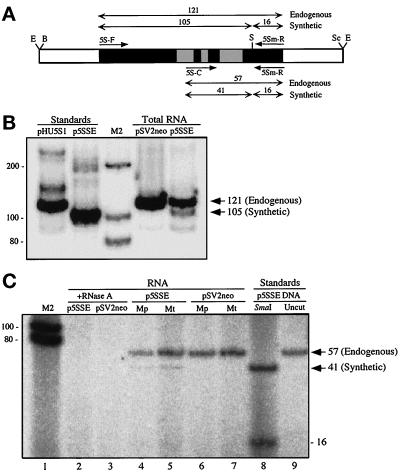
Figure 4.
Importation of synthetic 5S rRNA into human mitochondria. (A) Map of the insert of p5SSE. The two sets of primer pairs (short single-headed arrows) used to analyze the endogenous and synthetic variants and their corresponding SmaI-digested PCR products (double-headed arrows with the expected sizes above the arrows, in bp) are indicated. The 5S rRNA gene is in black with the known Pol III control elements (Willis, 1993) in the shaded boxes. The relevant restriction sites are indicated as follows: B, BamHI; E, EcoRI; S, SmaI; and Sc, SacI. (B) RT-PCR and RFLP analysis of total RNA from 293T cells that had been transfected transiently with p5SSE or pSV2neo, by the use of primers 5S-F and 5Sm-R, and electrophoresed through a 12% nondenaturing polyacrylamide gel. Note that the sizes of the SmaI digestions of RT-PCR products correspond exactly to the PCR products from pHU5S1 (wild-type) and p5SSE (A–C transversion) standards. Sizes of bands, in bp, are shown on the right. Sizes of M2 markers, in bp, are shown on the left. The 16-bp fragment is not resolved in this gel system. (C) RT-PCR and RFLP analysis of RNA isolated from highly purified mitochondria (Mt) and mitoplasts (Mp; obtained by the swell-contract method) from 293T cells transfected with p5SSE or pSV2neo, by the use of primers 5S-C and 5Sm-R, and electrophoresed through a 20% nondenaturing polyacrylamide gel. Note that the diagnostic 41-bp fragment is present in the p5SSE samples (lanes 4 and 5; the 16-bp fragments were diffuse and barely visible but were discernible in extremely dark exposures), with a size identical to that obtained in a PCR and RFLP from the p5SSE standard (lane 8), but is absent in the pSV2neo control samples (lanes 6 and 7). All RT-PCR species were abolished when mitochondria were treated with RNase A in the presence of SDS (lanes 2 and 3).