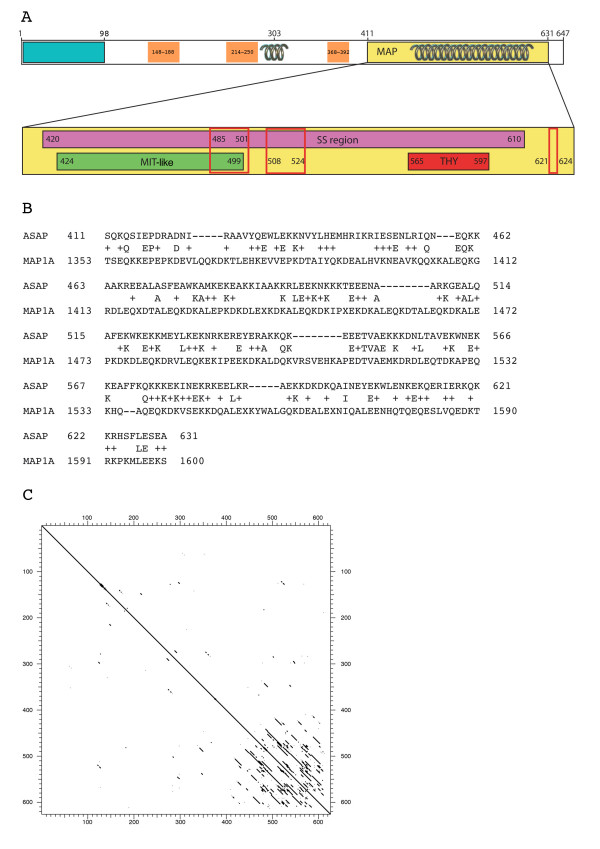
Figure 4.
Domains and motifs of the ASAP protein. (A) Top: General structure of ASAP. The residues are numbered 1 (N-terminal end) to 647 (C-terminal end). The MAP domain is indicated as well as the coiled-coil regions (springs). The blue rectangle indicates the region that is the most conserved during evolution in the N-terminal part (Fig. 2), orange boxes indicate moderatly conserved regions and uncoloured portions represent variable regions. Bottom: Detail of the MAP domain. This domain contains a self similarity (SS) region as MAP1A, an MIT-like and an THY domain. The nuclear localization signals (NLS) are boxed in red. MAP, microtubule-associated protein; MIT, microtubule interacting and trafficking domain; THY, thymosin beta actin-binding motif. (B) Comparison of ASAP with MAP1A. Blast comparison of ASAP with protein databases showing a similarity region with human MAP1A containing a high fraction of Glu and Lys (43, 21.8% and 52, 26.4% respectively in ASAP). (C) Protein self matrix of human ASAP. The comparison was performed using the Strider software (window 23 amino-acids, stringency 5 amino-acids). One region with self-similarity (SS-ASAP, amino-acids 420–610) corresponding to the region homologous to MAP1A, could be identified.