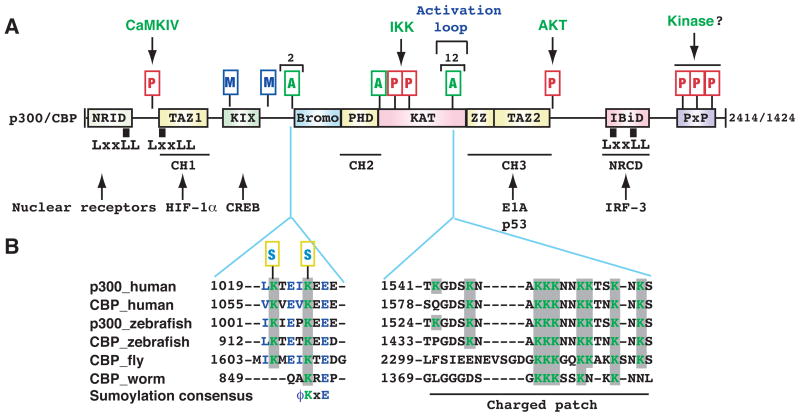
Figure 2. Autoacetylation and other modifications of p300 and CBP.
(A) Domain organization of human p300 and CBP. As mammalian p300 and CBP are functionally interchangeable in many in vitro assays, the term p300/CBP is used to refer to one or the other. Acetylation, phosphorylation, methylation and sumoylation sites are denoted with small rectangles containing the letters A, P, M and S, respectively. Within the activation loop of p300 and perhaps also CBP, there are 12 acetylation sites. The responsible kinases are indicated for the phosphorylation sites. Representative transcription factors that use p300 and CBP as coactivators are listed along with arrows pointing to the respective binding domains. Domain abbreviation: Bromo, bromodomain; CH1, Cys- & His-rich domain 1; IBiD, IRF-3 binding domain; KIX, kinase-inducing domain (KID) binding region; NRCD, nuclear receptor coactivator binding domain (synonymous to IBiD); NRID, nuclear receptor interacting domain; PxP, Pro-rich domain (no known function); TAZ1, transcriptional adaptor zinc finger; ZZ, zinc finger near the dystrophin WW domain.
(B) Regional sequence comparison of p300 and CBP from human and other species. The alignment was generated and shaded using ClustalW of MacVector 7.2 (Accelrys), using GenBank accession numbers Q09472, 119943104, 125830995, 125850731, 24640865, and 17552710. According to the new nomenclature system for histone-modifying enzymes (Allis et al., 2007), mammalian CBP and p300 are referred to as KAT3A and KAT3B, respectively. In the sumoylation consensus, φ is preferably a bulky residue (e.g., Leu, Ile or Val). Note that Lys 1057 of mouse CBP (1056 in humans) has been shown to be sumoylated (Kuo et al., 2005).