Abstract
Androgen receptor (AR) belongs to the nuclear receptor superfamily and mediates the biological actions of male sex steroids. In this work, we have characterized a novel 130-kDa Ser/Thr protein kinase ANPK that interacts with the zinc finger region of AR in vivo and in vitro. The catalytic kinase domain of ANPK shares considerable sequence similarity with the minibrain gene product, a protein kinase suggested to contribute to learning defects associated with Down syndrome. However, the rest of ANPK sequence, including the AR-interacting interface, exhibits no apparent homology with other proteins. ANPK is a nuclear protein that is widely expressed in mammalian tissues. Its overexpression enhances AR-dependent transcription in various cell lines. In addition to the zinc finger region, ligand-binding domain and activation function AF1 of AR are needed, as the activity of AR mutants devoid of these domains was not influenced by ANPK. The receptor protein does not appear to be a substrate for ANPK in vitro, and overexpression of ANPK does not increase the extent of AR phosphorylation in vivo. In view of this, it is likely that ANPK-mediated activation of AR function is exerted through modification of AR-associated proteins, such as coregulatory factors, and/or through stabilization of the receptor protein against degradation.
INTRODUCTION
The androgen receptor (AR) mediates the biological actions of physiological androgens (Quigley et al., 1995). DNA-bound transcription factors, such as AR and other nuclear receptors, have been postulated to stimulate the efficiency of transcription by affecting directly or indirectly the assembly of basal transcription factors into the preinitiation complex, thereby increasing the rate of transcription initiation (Tjian and Maniatis, 1994; Horwitz et al., 1996; Beato and Sanchez-Pacheco, 1996). However, regulation of receptor function also involves cross-talk with other signaling pathways and interactions with other transcription factors and coregulatory proteins (Beato et al., 1995). Although nuclear receptors may contact directly some members of the basal transcription machinery (Ing et al., 1992; Blanco et al., 1995; Hadzig et al., 1995; Schulman et al., 1995; McEwan and Gustafsson, 1997) or TATA-binding protein–associated factors (Jacq et al., 1994; Schwerk et al., 1995; Mengus et al., 1997), they appear to employ preferentially coregulators to interact with the transcription machinery (Beato and Sanchez-Pacheco 1996; Horwitz et al., 1996). Examples of the coregulators include RIP-140 (Cavaillés et al., 1995), TIF1 (Le Douarin et al., 1995), TRIP1/SUG1 (Lee et al., 1995; vom Baur et al., 1996), ARA70 (Yeh and Chang, 1996), CBP/p300 (Chakravarti et al., 1996; Hanstein et al., 1996; Kamei et al., 1996), and SRC-1 (Oñate et al., 1995) and its variants or related proteins, such as GRIP1, TIF2, AIB1, and TRAM-1 (Hong et al., 1996; Voegel et al., 1996; Anzick et al., 1997; Takeshita et al., 1997). Most of the these coregulators have been discovered through protein–protein interaction screening techniques using the ligand-binding domains (LBDs) of nuclear receptors as target sequences.
Even though the zinc finger region (ZFR) of steroid receptors is mainly involved in DNA binding and homodimerization, recent evidence suggests that it also plays an important role in mediating protein–protein interactions with other regulatory proteins (Schena et al., 1989; Beato et al., 1995; Starr et al., 1996; Lichtarge et al., 1997; Moilanen et al., 1998). In our search for proteins capable of interacting with AR ZFR, we have identified a nuclear protein kinase, which we termed ANPK for androgen receptor-interacting nuclear protein kinase. The catalytic kinase domain of ANPK shares marked similarity with that of the minibrain gene product, a protein kinase recently implicated in learning defects associated with Down syndrome (Smith et al., 1997), whereas the C-terminal half of this 130-kDa protein exhibits no apparent sequence homology with other proteins. ANPK is a functionally active protein kinase. It interacts with AR in a hormone-dependent manner and activates AR-mediated transcription, suggesting that ANPK acts as a coregulator of AR function.
MATERIALS AND METHODS
Materials
pPB(−285/+32)-LUC is a reporter that contains nucleotides (nt) −285 to +32 of the rat probasin promoter (Palvimo et al., 1996) and pGRE2-E1b-CAT (pARE2-E1b-CAT in this report) contains two copies of rat tyrosine aminotransferase glucocorticoid/progesterone/androgen response element (GRE/PRE/ARE) inserted upstream of the adenovirus E1b TATA sequence (a gift from Dr. J. Cidlowski, NIEHS, Research Triangle Park, NC) (Allgood et al., 1993). pSG5-hPR1 and pHG0 encoding human PR1 and GR, respectively, were gifts from Dr. Pierre Chambon (INSERM, Illkirch, France). pSG5-hGR was created by inserting hGR coding sequence from pGH0 as a BamHI fragment into the BamHI site of pSG5 (Stratagene, La Jolla, CA). pCB6-WT18A (WT1) encoding Wilms’ tumor gene product was from Dr. Frank J. Rauscher III (Wistar Institute, Philadelphia, PA). pMOR encoding mouse ER was a gift from Dr. Malcolm G. Parker (Imperial Cancer Research Fund, London, UK). pG5-CAT contains five Gal4-binding sites in front of the adenovirus E1b minimal promoter driving the CAT gene (CLONTECH, Palo Alto, CA). The β-galactosidase (β-gal) expression plasmid pCMVβ was purchased from CLONTECH. The following mammalian two-hybrid system vectors were used (all from CLONTECH): pM for expressing DBD of the Saccharomyces cerevisiae Gal4 protein (residues 1–147), pVP16 for expressing the transcriptional activation domain (VP16 AD) of the herpes simplex virus VP16 protein (amino acid residues 411–456), pM-VP16 encoding Gal4 DBD-VP16 AD fusion protein and pVP16-CP for expressing a fusion of VP16 AD to the polyoma virus coat protein. Testosterone was purchased from Makor Chemicals (Jerusalem, Israel), and progesterone, dexamethasone, and rabbit myelin basic protein (MBP) were from Sigma Chemical (St. Louis, MO). [3H]Acetyl-CoA was purchased from Du Pont-New England Nuclear (Boston, MA). Luciferase assay reagent, TNT T7-coupled rabbit reticulocyte system and recombinant human CDC2 kinase complexed with cyclin B were purchased from Promega (Madison, WI). Recombinant rat ERK2 and PHAS-I substrate were from Stratagene. PhosphoSpots cellulose strips containing covalently bound peptide substrates for protein kinases were purchased from Jerini Bio Tools (Berlin, Germany). The yeast two-hybrid system vectors, pVP16 and pLex-a and pLexN-a, based on pBTM116 (Bartel et al., 1993) and encoding bacterial LexA and LexA fused to a SV40 large T antigen nuclear localization signal N-terminal to the polylinker, respectively, were kind gifts from Dr. Stanley Hollenberg (Vollum Institute, Oregon Health Sciences University, Portland, OR). Recombinant c-Jun was expressed and affinity purified as described by Kallio et al. (1995). Histone H1 and HMG14 were purified from calf thymus (Palvimo and Mäenpää, 1988), and core histones were isolated from rat thymus. Histone H3 was purchased from Boehringer Mannheim (Indianapolis, IN), and [γ-32P]ATP (3 Ci/mmol) was from Amersham Life Science (Arlington Heights, IL).
Isolation of Partial cDNAs for AR-interacting Proteins Using a Yeast Two-Hybrid System
Yeast two-hybrid screening of mouse embryo E10.5 cDNA library (a gift from S. Hollenberg) was used to identify clones that interact with human AR (hAR) ZFR as described by Hollenberg et al. (1995). LexA fusion expression vector pLex-ZFR (residues 554–644), which uses LexA as a DNA-binding component, was generated by inserting a PCR amplified cDNA fragment encoding residues 554–644 of hAR in-frame into the BamHI/SalI site of pLex-a and used as a bait. The library with random-primed size-selected cDNA inserts (∼500 nt) in pVP16 vector uses a region of herpex simplex virus VP16 as a transcriptional activator. The yeast strain L40 (MATa trp1–901 leu 2–3, 112 LYS::(lexAop)4-HIS3 URA3:: (lexAop)8-LacZ) was used in the assay. Approximately 2.5 × 107 transformants were screened for interaction in the presence of 0.5 mM 3-aminotriazole. All clones potentially interacting with AR ZFR were cured of the bait plasmid and tested against the negative control plasmids, pLex-a, pLex-Lamin, and pLex-WT1ZF (WT1ZF; Wilms tumor protein zinc finger residues 312–419) by a mating strategy using AMR70 (MATa his3Δ200 lys2–801am trp1–901 leu 2–3, 112 URA3::(lexAop)8-LacZ) (Hollenberg et al., 1995). Five of the 28 positive clones contained 400–500 nt cDNAs corresponding to overlapping fragments of ANPK. Liquid β-galactosidase assay was performed as described previously (Moilanen et al., 1997).
cDNA Cloning and Characterization
A rat testis λZapII cDNA library was screened with 32P-labeled ANPK cDNA corresponding to amino acids 766–920 (ANPK ID) using standard hybridization and high-stringency washing conditions (Ausubel et al., 1997). Positive clones were converted in vivo into pBluescript plasmids according to manufacturer’s instructions. The BLAST program (Altschul et al., 1990) was used to search for DNA and protein sequence homologies in the databases at National Center for Biotechnology Information.
Plasmid Construction
The rat (r) AR expression vectors, pSGrAR, rARΔ46–408, rARΔ641–902, and rARΔ46–408/Δ641–902, were constructed as previously described (Palvimo et al., 1993; Ikonen et al., 1997), and the hAR expression vector pSG5hAR was constructed as described by Adeyemo et al. (1993). Fusion vectors containing indicated (in parentheses) amino acids of AR were constructed as follows: LexA fusion expression vectors, pLex-ZFR-s (residues 554–628), pLex-LBD (residues 657–919), and pLex-HLBD (residues 624–919), were generated by inserting PCR-amplified cDNA fragments of hAR in-frame into the BamHI/SalI site of pLex-a (pLexN-a for ZFR-s). The pLex-rAR and pM-rAR coding for full-length rAR-LexA fusions was constructed by transferring rAR corresponding to the BamHI/PstI fragment from pGEM-3Z-rAR (Moilanen et al., 1997) into pLex-a and pM. pLex-WT1-ZF (zinc finger, residues 312–419) was created by inserting a PCR-amplified cDNA fragment into the BamHI/SalI site of pLex-a. Glutathione S-transferase (GST) fusion expression vector for hAR-ZFR has been described previously (Kallio et al., 1995).
To construct pFLAG-ANPK(2–1191) and pFLAG-ANPK(159–1191), PCR-amplified fragments of ANPK cDNA corresponding to amino acids 2–336 and 159–336, respectively, with KpnI sites in the upstream primers, were digested with KpnI/BamHI and cloned into the corresponding site of pFLAG-CMV-2 (Kodak IBI, Rochester, NY), and a cDNA fragment encoding amino acids 337–1191 was subsequently inserted downstream of the BamHI site. FLAG-ANPK(K226R) mutant containing Lys226 converted to Arg in the putative ATP-binding site was constructed as pFLAG-ANPK, but the fragment corresponding to amino acids 159–336 was generated by the overlapping PCR mutagenesis strategy (Ausubel et al., 1997). ANPK(S357A/Y359A) mutant having both Ser357 and Tyr359 converted to Ala was created as above, but the overlapping PCR mutagenesis strategy was used to generate a cDNA fragment corresponding to amino acids 159–644. pFLAG-ANPK(159–772) was made by replacing a SmaI fragment of ANPK corresponding to amino acids 417–1191 with a PCR-generated cDNA fragment encoding amino acids 417–789. pGEX-ANPK and pGEX-ANPK-ID were obtained by cloning PCR-generated cDNA fragments corresponding to ANPK(159–920) and ANPK(766–920) into the EcoRI/SalI or BamHI/SalI site of pGEX-5X-1 expression vector (Pharmacia, Piscataway, NJ), respectively. The mammalian two-hybrid expression vector pVP16-ANPK was created by cloning the ANPK(159–920) fragment from pGEX-ANPK into the EcoRI/SalI site in pVP16 vector. All constructs were verified by DNA sequencing using the ALFExpress system (Pharmacia Biotech).
Cell Culture, Transfections, Metabolic Labeling with [32P]Orthophosphate, and Ligand-binding Assays
All mammalian cell lines, except for S115 cells (from Dr. P. Härkönen, University of Turku, Turku, Finland), were obtained from American Type Culture Collection (ATCC, Rockville, MD) and maintained according to ATCC’s instructions. S115 were cultured as described by Palvimo et al. (1996). Cells were transfected using the calcium phosphate precipitation method as described previously (Palvimo et al., 1993; Ikonen et al., 1997). The cells (1.5 × 106) were plated on a 10-cm dish 24 h before addition of the precipitate with indicated amounts of expression and reporter vectors. β-Gal expression plasmid pCMVβ (2 μg/10-cm plate) was used as an internal control for transfection efficiency. For domain interaction studies (the mammalian two-hybrid system), CV-1 cells were transfected using DOTAP reagent as described by Ikonen et al. (1997). Eighteen hours after transfection, the medium was changed to one containing charcoal-stripped 2% (vol/vol) FBS in the presence or absence of testosterone as depicted in the figure legends. CAT and β-galactosidase activities were assayed as previously described (Eastman, 1987; Rosenthal, 1987; Palvimo et al., 1993). For metabolic labeling with [32P]orthophosphate, CV-1 cells on 10-cm dishes were transfected with 5 μg of pSG5-rAR (or empty pSG5) along with 15 μg of pFLAG-ANPK(2–1191) (or empty pFLAG-CMV2). After 48 h, the cells were washed twice with phosphate-free medium and incubated for 2.5 h in the presence of 0.25 mCi [32P]orthophosphate in phosphate-free medium supplemented with 100 nM testosterone and 2% (vol/vol) FBS (Zhou et al. 1995). Protein concentration was determined using Bio-Rad (Richmond, CA) reagents according to the manufacturer’s instructions. Luciferase activity was determined with reagents from Promega using a Luminoskan RT reader (Labsystems, Helsinki, Finland) (Palvimo et al., 1996). Whole cell steroid-binding assays were performed as described by Palvimo et al. (1993). Statistical analyses of the data were carried out using two-tailed Student’s t test.
Proteins, Antibodies, and Immunoblotting
Histidine-tagged H6-ZFR (residues 554–644 of hAR) and H6-N-TERM (residues 141–547) and GST-HLBD (residues 624–919) were expressed in bacteria and purified by affinity chromatography (Palvimo et al., 1996). Bacterially expressed GST-ANPK(159–920) and GST-ANPK(766–920) were purified by affinity chromatography as described (Kallio et al., 1995), except that fusion proteins were eluted in a buffer containing PBS (140 mM NaCl, 20 mM Na-phosphate, pH 7.4), 10% (vol/vol) glycerol, and 10 mM reduced glutathione. Polyclonal antisera were raised against purified GST-ANPK(766–920) in rabbits using 50 μg of protein at each immunization.
Whole cell extracts were prepared as previously described (Ikonen et al., 1994). For immunoblot analysis, 15 μg of cell extracts were fractionated by electrophoresis on polyacrylamide gels under denaturing conditions (Laemmli, 1970) and electroblotted onto Immobilon-P membrane (Millipore, Bedford, MA) or Hybond ECL membrane (Amersham). Immunocomplexes were visualized using either ECL Western blotting detection reagents from Amersham or 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium substrate system from Zymed according to the manufacturers’ instructions.
Protein–Protein Interaction in Vitro
Protein–protein affinity chromatography using purified GST-AR ZFR (Kallio et al., 1995) or GST alone bound to Glutathione Sepharose (5 μg protein/40 μl resin) and 10 μl [35S]methionine-labeled in vitro- translated ANPK was carried out in a buffer containing 50 mM Tris-Cl (pH 7.8), 150 mM KCl, 0.1% (vol/vol) Nonidet-40, 0.1% (vol/vol) Triton X-100, 5 mM MgCl2, 0.5 mM EDTA, 10% (vol/vol) glycerol, 50 μM ZnCl2, 0.1 mM Na-orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 10 μg/ml aprotinin, and 5 μg/ml pepstatin-A in a total volume of 500 μl at 4°C for overnight. The resin was subsequently washed four times with 1 ml of binding buffer. Bound proteins were released in the Laemmli sample buffer (Laemmli, 1970), subjected to electrophoresis under denaturing conditions, and visualized by fluorography.
Immunocytofluorescence and Immunohistochemistry
CV-1 cells seeded on glass cover slips on 10-cm plastic plates were transfected using DOTAP reagent with 1 μg of pFLAG-ANPK and 9 μg of pBSIISK (Stratagene) as carrier DNA. Cells were fixed in 4% (wt/vol) paraformaldehyde in PBS and permeabilized with Triton X-100, and the ANPK protein was visualized using anti-FLAG M2 monoclonal antibody (Kodak IBI, 1:50 dilution) or anti-ANPK antiserum (1:500 dilution) and FITC-conjugated goat anti-mouse or anti-rabbit secondary antibody, respectively (1:200 dilution, Jackson ImmunoResearch Laboratories, West Grove, PA) or, in double immunofluorescence labeling for confocal microscopy, using lissamine–rhodamine-conjugated goat anti-mouse secondary antibody. AR was visualized with a polyclonal rabbit antiserum raised against full-length rAR (K183) and FITC-conjugated goat anti-rabbit secondary antibody as described previously (Karvonen et al., 1997).
Rat prostates were frozen on dry ice and stored at −80°C. Seven-micrometer cryostat sections from Tissue-Tek (Miles, Kankakee, IL)-embedded tissues were fixed in acetone at −20°C and stored at −20°C. After rehydration in PBS, sections were incubated with 0.03% (vol/vol) hydrogen peroxide in methanol for 30 min at room temperature to reduce endogenous peroxidase activity and washed extensively in PBS. Nonspecific binding was blocked by incubating tissue sections for 30 min in 10% normal rabbit serum in PBS or in 1% blocking reagent (DIG DNA labeling and detection Kit, Boehringer Mannheim) in 100 mM Tris-Cl and 150 mM NaCl (pH 7.9). Polyclonal ANPK antiserum was used in a 1:1000 dilution and incubated at room temperature for 1 h or at 4°C for 14–16 h. After washing with PBS, biotin-labeled anti-rabbit-IgG and AB-complex from Vectastain Elite-Kit (Vector Laboratories, Burlingame, CA) were applied following the manufacturer’s instructions. Peroxidase reaction was carried out with 0.02% (wt/vol) 3-amino-9-ethylcarbazole in 50 mM Na-acetate (pH 5.0) for 20 min at room temperature.
Immunoprecipitation, Kinase Assays, and Phosphoamino Acid Analysis
Expression plasmids for wild-type or mutant FLAG-tagged ANPK were used to transfect CHO cells using the calcium phosphate method. Two days after transfection, the medium was removed and cell monolayers washed twice with cold PBS. For immunoprecipitation, cells were lysed in ice-cold buffer containing 25 mM HEPES (pH 7.5), 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 1 mM dithiothreitol, 20 mM β-glycerophosphate, 40 mM p-nitrophenyl phosphate, 0.1 mM Na3VO4, 0.5 μM okadaic acid, 0.5% Triton X-100, and the following protease inhibitors: 0.5 mM PMSF, 5 μg/ml leupeptin, 5 μg/ml pepstatin A, and 10 μg/ml aprotinin. The collected cell debris and lysate were clarified by centrifugation for 15 min at 16,000 × g at 4°C and subjected to preclearing with normal rabbit serum and GammaBind G Sepharose or Protein A Sepharose (Pharmacia Biotech). AR from [32P]orthophosphate-labeled cells was immunoprecipitated with K183 antiserum using Protein A Sepharose and washed three times with lysis buffer. ANPK was precipitated by incubating lysate with M2 monoclonal antibody (Kodak IBI) (directed against the FLAG epitope) and GammaBind G Sepharose. For immune kinase complex assays, immunoprecipitates were washed three times with lysis buffer and twice with kinase buffer containing 25 mM HEPES (pH 7.5), 10 mM MgCl2, 0.1 mM Na3VO4, 20 mM β-glycerophosphate, and 1 mM dithiothreitol. For catalytic kinase assay, immunoprecipitation pellets or bacterially expressed GST-ANPK(159–920) were incubated with indicated concentrations of substrates and 35 μM [γ-32P]ATP in 20 μl of kinase buffer. The kinase reaction was performed at 30°C for 30 min and terminated with 20 μl of 2× Laemmli sample buffer, and the products were resolved by PAGE under denaturing conditions.
Phosphorylated proteins obtained from the immune complex kinase assays were transferred electrophoretically to PVDF membranes. The bands containing phosphoproteins were excised and hydrolyzed in 6 M HCl at 105°C for 75 min. The supernatants were lyophilized and dissolved in 10 μl of H2O containing cold phosphoserine, phosphothreonine, and phosphotyrosine (0.1 μmol each) as markers. Phosphoamino acids were resolved by electrophoresis (30 mA, 45 min) at pH 2.5 (Jelinek and Weber, 1993).
RNA Isolation and Northern Blot Analysis
RNA was extracted from adult rat tissues by the LiCl-urea precipitation method (Auffray and Rougeon, 1980) and enriched for poly(A)+ RNA by oligo(dT)-cellulose chromatography. Polyadenylated RNA samples (5 μg/lane) were fractionated on 1.3% agarose gels containing 2.2 M formaldehyde, transferred to Hybond-N nylon membrane (Amersham), and immobilized onto the membrane by exposure to UV light (Stratalinker, Stratagene). Membrane was hybridized to 32P-labeled ANPK cDNA, washed at high stringency (0.2× SSC, 0.1% SDS, 52°C), and subjected to autoradiography. Human multiple tissue Northern blot (CLONTECH) was hybridized according to manufacturer’s protocol, washed at high stringency (0.1× SSC, 0.1% SDS, 35°C), and subjected to autoradiography.
RESULTS
Isolation of cDNAs for ANPK
The yeast two-hybrid system was used to identify proteins that interact with the ZFR of hAR. A fusion between LexA protein and ZFR, including part of the hinge region (LexA-ZFR, amino acids 554–644 of hAR), was used as a bait to screen a mouse cDNA library fused to VP16 activation domain (VP16 AD) (Hollenberg et al., 1995). Five of the positive clones that interacted specifically and reproducibly with AR ZFR shared overlapping cDNA sequences of a previously uncharacterized protein, termed ANPK in this report.
A rat testis cDNA library was screened to isolate longer cDNA clones than those in the initial screening of the 10.5-d mouse embryo cDNA library (Hollenberg et al., 1995). The longest clone of 4120 nt in size contained an open reading frame for 1191 amino acid residues with a predicted molecular size of 130 kDa for the translated protein (Figure 1A). The cDNA clone included 471 nt of 5′-untranslated region (UTR) that was GC-rich and 74 nt of 3′-UTR containing a potential polyadenylation signal (AAUAAA). There was an in-frame stop codon 9 nt upstream of the first AUG, suggesting that the cDNA clone was likely to encode a full-length protein.
Figure 1.
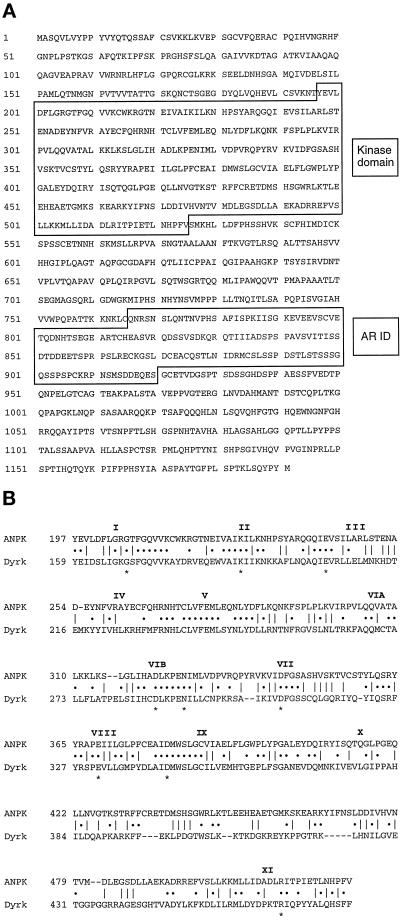
Primary structure of rat ANPK and sequence comparison. (A) Predicted amino acid sequence of ANPK (GenBank accession no. AF036959). The numbers on the left depict amino acid positions. The catalytic kinase domain and the AR interaction domain (AR ID) are boxed. (B) Sequence alignment of protein kinase domains of ANPK and Dyrk. Roman numerals on the top denote the conserved kinase subdomains according to Hanks and Quinn (1991). Invariant residues in all the protein kinases are depicted with asterisks below the residues.
The N-terminal region of ANPK (residues 197–526) showed extensive sequence homology to many protein kinases, and all of the 12 subdomains common to known protein kinases were conserved within ANPK (Hanks and Quinn, 1991; Hanks and Hunter, 1995) (Figure 1B). Sequence alignments also predicted that ANPK is specific for Ser and Thr residues, as sequences of subdomains VIB and VIII matched with a Ser/Thr-specific but not a Tyr-specific consensus sequence (Figure 1B). However, whether ANPK represents a new dual specificity kinase cannot be predicted on the basis of primary sequence alone (Hanks and Hunter, 1995). Comparison of the kinase domain sequence of ANPK with those available in current data resources indicated that it has greatest similarity to protein kinases of the Clk subfamily (Hanks and Quinn, 1991; Hanks and Hunter, 1995) but is not a counterpart of any particular member. Among the characterized protein kinases, ANPK shares highest similarity with the kinase domain of Yak1 (Garrett and Broach, 1989) from yeast and that of Drosophila minibrain (Tejedor et al., 1995) or its human and rat homologs, MNB and Dyrk, respectively (Kentrup et al., 1996; Shindoh et al., 1996). ANPK and Dyrk are 39% identical throughout the kinase domain (Figure 1B), whereas the degree of identity between ANPK and Yak1 catalytic domains is 43%, and ANPK and Clk1 share 32% identity. Also C. elegans chromosome III appears to encode a protein kinase whose catalytic domain shares 63% identity with that of ANPK (Wilson et al., 1994). The extreme N-terminal region and the C-terminal half of ANPK seem unique, as sequences outside the kinase domain do not contain any known functional motifs or share homology with previously characterized proteins.
Expression and Localization of ANPK
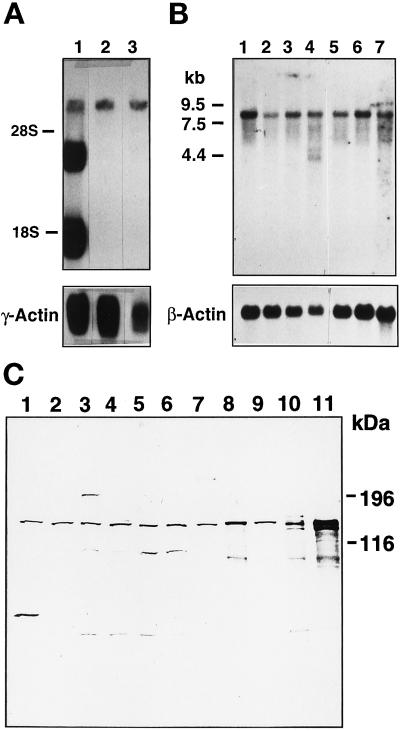
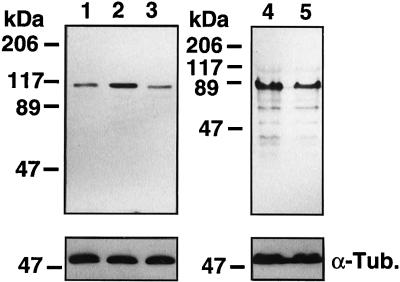
RNA blot analyses revealed expression of ∼7 kilobase (kb) ANPK mRNA in many rat tissues (Figure 2A). Two additional smaller ANPK mRNA species, ∼4 kb and 2 kb in size, were expressed to very high levels in rat testis. It is currently unknown whether these smaller mRNA species originate from alternative splicing of the primary transcript or the use of a testis-specific promoter. An 8-kb ANPK mRNA was detected in various human tissues; in addition, human testis contained smaller ANPK mRNA species, even though their relative abundance was not as high as in rat testis (Figure 2B). Dot blot analysis revealed the highest levels of ANPK mRNA in human skeletal muscle and heart (our unpublished data). The level of ANPK protein was examined by immunoblot analysis of lysates from several cell lines using a rabbit antiserum raised against bacterially expressed GST-ANPK(766–920). ANPK appeared to be widely expressed, and a ∼160-kDa protein comigrating with ANPK produced by translation in vitro was detected in all cell lines examined, including CV-1 cells (Figure 2C).
Figure 2.
Expression of ANPK. (A) Northern blot of RNA from rat tissues. Polyadenylated RNA samples (5 μg/lane) from testis (1), prostate (2), and brain (3). (B) Northern blot of poly(A)+ RNA (2 μg/lane) from various human tissues. Human multiple tissue Northern blot II (CLONTECH) contains RNA from spleen (1), thymus (2), prostate (3), testis (4), small intestine (5), colon (6), and peripheral blood leukocytes (7). Both blots were hybridized with 32P-labeled ANPK cDNA as described in MATERIALS AND METHODS. To check the integrity of the RNA samples, the rat blot was reprobed with rat γ-actin and the human blot with human β-actin cDNA, respectively. (C) Expression of ANPK protein in various cell lines. Fifteen micrograms of whole cell extracts from PC12 (1), N2A (2), LNCaP (3), CHO (4, 5), PC-3 (6), CV-1 (7), and COS-1 cells transfected with an ANPK expression vector (8), and mock-transfected COS-1 cells (9), or 2 μl of ANPK(1–1191) (10) and ANPK(159–1191) (11) produced by translation in vitro using reticulocyte lysate system were separated on 7.5% polyacrylamide gel under denaturing conditions and immunoblotted using rabbit antiserum against GST-ANPK(766–920).
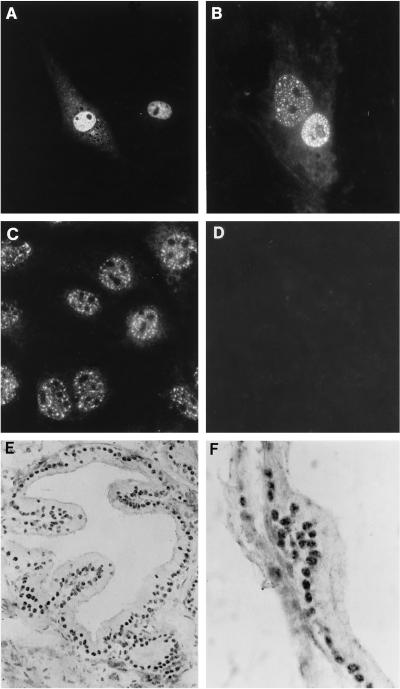
Subcellular localization of ANPK was initially assessed in cultured cells by indirect immunofluoresence using affinity-purified anti-ANPK antibodies. Even though no nuclear localization signal was apparent in the ANPK sequence, transiently expressed ANPK resided almost exclusively in nuclei of CV-1 cells and displayed a speckled pattern of distribution (Figure 3, A and B). Additionally, the endogenous nuclear ANPK of mouse S115 cells, which also express AR protein, exhibited a similar punctate pattern (Figure 3C). In rat prostate, ANPK antigen was detected in nuclei of secretory epithelial cells that exhibited a granular staining pattern with a few ANPK-positive granulae per nucleus (Figure 3, E and F).
Figure 3.
Localization of ANPK in cultured cells and prostate. (A and B) CV-1 cells seeded on glass coverslips on 10-cm plastic plates were transfected with 1 μg of FLAG-ANPK(2–1191) expression vector using DOTAP transfection reagent as described in MATERIALS AND METHODS. Cells were fixed in 4% (wt/vol) paraformaldehyde, permeabilized, and ANPK antigen was visualized using (A) affinity-purified rabbit antiserum raised against GST-ANPK(766–920) or (B) anti-FLAG M2 antibody. (C) The distribution of endogenous ANPK in S115 cells as determined by using anti-ANPK antiserum. (D) Immunofluorescence of S115 cells with anti-ANPK antiserum neutralized with purified GST-ANPK(766–920) fusion protein. (E and F) Distribution of ANPK in rat prostate. Immunoperoxidase technique was applied to visualize ANPK antigen using rabbit antiserum raised against GST-ANPK(766–920). (E) ANPK is localized in nuclei of epithelial cells. (F) A higher magnification showing granular clusters of ANPK immunoreactivity in epithelial cell nuclei. Original magnification; E, ×760; F, ×1,900.
Protein Kinase Activity of ANPK
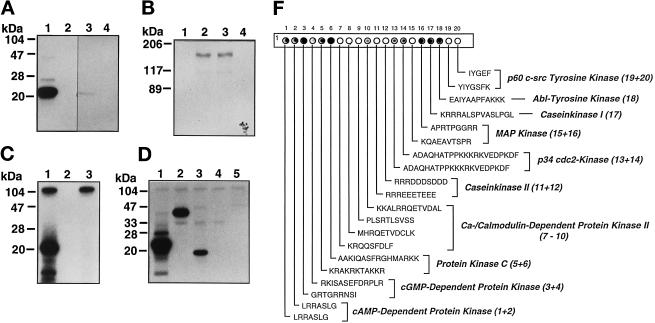
To verify that ANPK was indeed a protein kinase, FLAG-tagged ANPK(159–1191) produced in CHO cells was subjected to immune complex kinase assays using myelin basic protein (MBP) as the substrate. Immunopurified ANPK(159–1191) efficiently phosphorylated MBP (Figure 4A, lane 1), whereas no incorporation of 32P into MBP was observed in the absence of kinase (lane 2). As a control, we expressed ANPK(K226R) in which Lys226 in the consensus ATP-binding site was converted to Arg. ANPK(K226R) was catalytically inactive, indicating that the ANPK kinase activity was not due to coimmunoprecipitated or contaminating proteins (Figure 4A, lane 3). Similar to Yak1 or Dyrk (Garrett et al., 1991; Kentrup et al., 1996), ANPK was autophosphorylated (Figure 4B, lanes 2 and 3), whereas ATP-binding–deficient ANPK(K226R) failed to show autophosphorylation (Figure 4B, lanes 1 and 4). ANPK was also autophosphorylated in vivo (our unpublished data). Additionally, residues 159–920 of ANPK expressed as a GST fusion protein in E. coli underwent autophosphorylation and behaved as an active MBP-phosphorylating kinase in vitro (Figure 4C).
Figure 4.
Protein kinase activity of ANPK. CHO cells were transiently transfected with 10 μg of expression vectors for FLAG-tagged ANPK(159–1191) and ANPK(K226R), in which Lys226 in the ATP-binding site was converted to an Arg. Proteins were immunoprecipitated using anti-FLAG mAb and subjected to immune complex kinase assays. (A) Phosphorylation of myelin basic protein (MBP, 5 μM) by immunopurified ANPK(159–1191) (lane 1), but not by ANPK(K226R) (lane 3), ANPK(159–1191), and ANPK(K226R) in the absence of MBP (lanes 2 and 4, respectively). (B) Autophosphorylation by ANPK(159–1191) (lanes 2 and 3) and ANPK(K226R) (lanes 1 and 4). (C) Protein kinase activity of purified GST-ANPK(159–920) expressed in E. coli. GST-ANPK(159–920) with MBP (lane 1), autophosphorylation of GST-ANPK(159–920) in the absence of MBP (lane 3), and MBP in the absence of GST-ANPK(159–920) (lane 2). (D) Comparison of MBP (lane 1), c-Jun (lane 2), histone H3 (lane 3), histone H1 (lane 4), and GST-ANPK(802–920) (lane 5) as substrates for GST-ANPK(159–920). Reaction mixture (20 μl) contained 0.7 μg of GST-ANPK(159–920) and 5 μM substrate. After 30°C for 30 min, the reactions were terminated by addition of 20 μl of 2× Laemmli sample buffer, after which the products were subjected to electrophoresis under denaturing conditions on 15% (A, C, and D) or 7.5% polyacrylamide gels (B) and visualized by autoradiography. (F) Solid-phase phosphorylation of synthetic peptides by ANPK. PhosphoSpots test strip (Jerini Bio Tools) containing covalently bound substrate peptides for indicated kinases was phosphorylated by incubating with 4 μg of GST-ANPK(159–920) in 500 μl of 2× kinase buffer containing 100 μM [γ-32P]ATP at 22°C for 20 min. The reaction was stopped and filter washed extensively according to manufacturer’s instructions and analyzed by autoradiography. (The peptide no. 19 contains both Ser and Tyr residues as potential phosphoacceptor sites.)
To characterize further the kinase activity of ANPK, histones, a nonhistone protein HMG14, and c-Jun were used as model substrates under cell-free conditions. GST-ANPK was able to phosphorylate c-Jun and histone H3, albeit less efficiently than MBP (Figure 4D). Other core histones, linker histone H1, and HMG14, which are all very basic proteins and good in vitro substrates for many Ser/Thr protein kinases, were not modified by ANPK (Figure 4D and our unpublished data). Solid-phase phosphorylation of a set of recognition sequence peptides for various well-characterized protein kinases revealed that ANPK is not a proline-directed kinase, as could have been anticipated on the basis of efficient phosphorylation of MBP (Figure 4F). ANPK prefers phosphoacceptor sites in the vicinity of or, especially, surrounded by basic amino acids: protein kinase C recognition peptide AAKIQASFRGHMARKK served as the most efficient substrate. Ser or Thr residues embedded in an acidic surrounding, such as peptides RRREEETEEE and RRRDDDSDDD, were very poor substrates. Interestingly, ANPK appears to be capable of transferring phosphate also to Tyr residues in basic peptides, as EAIYAAPFAKKK peptide was phosphorylated. Taken together, these data indicate that the substrate specificity of ANPK differs from those of previously characterized cyclin-dependent kinases (cdks) and MAP kinases.
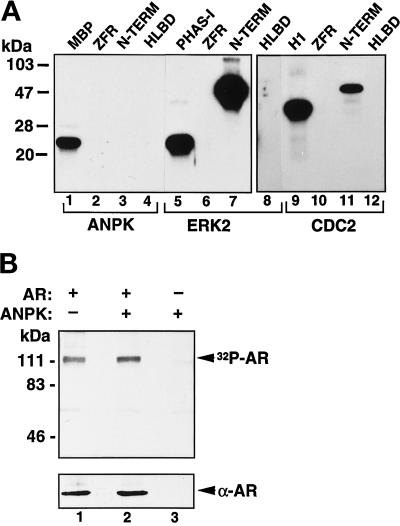
The major phosphorylation sites of AR appear to be proline-directed, but the kinase(s) phosphorylating them is (are) unknown (Zhou et al., 1995; Blok et al., 1998). Even though the above results do not imply that ANPK prefers phosphoacceptor sites located close to prolines, AR was still considered as a potential ANPK substrate. However, neither immunopurified FLAG-ANPK nor recombinant GST-ANPK phosphorylated AR in vitro independent of the receptor preparation used as a substrate, including full-length AR expressed and purified from insect cells (Kallio et al., 1995), and bacterially expressed and purified H6- or GST- fusion proteins corresponding to various functional domains of AR (Figure 5A and our unpublished data). Nevertheless, H6-AR fusion protein containing the receptor N-terminal residues 141–547, which encompass the transcription activation function AF1, served as an excellent in vitro substrate for extracellular signal-regulated kinase 2 (ERK2) with an efficiency better than the established ERK2 substrate PHAS-I (Figure 5A, lanes 5 and 7). Also cyclin B-activated CDC2 kinase phosphorylated the N-terminal AR domain (lane 11). In keeping with the above data, the extent of phosphorylation of full-length AR protein was not altered by coexpression of ANPK CV-1 cells, as judged by 32P incorporation into immunoprecipitated AR (Figure 5B). Collectively, these results argue against AR being a substrate for ANPK.
Figure 5.
Phosphorylation of AR in vitro and in vivo. (A) AR is phosphorylated in vitro by ERK2 and CDC2 kinase, but not by ANPK. Histidine tagged H6-ZFR (residues 554–644 of hAR), H6-N-TERM (residues 141–547), and GST-HLBD (residues 624–919) were expressed in bacteria, affinity purified, and phosphorylated using indicated protein kinases under the conditions described in Figure 4, except that the concentration of GST-HLBD was 1 μM. ERK2 and CDC2 were used at 5 ng/μl and 0.25 U/μl, respectively. PHAS-I and histone H1 served as established control substrates for ERK2 and CDC2, respectively. (B) Effect of cotransfected ANPK on phosphorylation of AR in vivo. CV-1 cells were cotransfected by the calcium phosphate method with expression vectors for AR and ANPK as indicated. After a 48-h culture in the presence of 100 nM testosterone, cells were metabolically labeled for 2.5 h with [32P]orthophosphate, and AR was immunoprecipitated as described in MATERIALS AND METHODS. In vivo labeled AR was subjected to electrophoresis under denaturing conditions on 7.5% polyacrylamide gel and visualized by autoradiography. Lower panel shows immunoblot analysis of the same samples using rabbit K333 antiserum raised against full-length rAR.
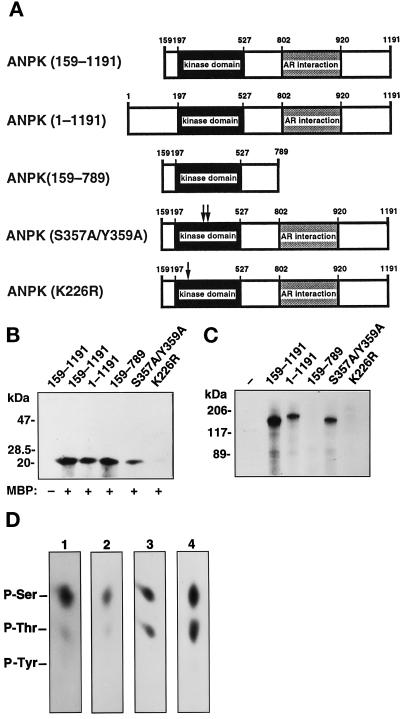
A series of FLAG-tagged ANPK mutants (Figure 6A) was generated to examine the importance of different regions of ANPK for its kinase activity. Full-length ANPK phosphorylated MBP approximately to the same extent as N-terminally truncated ANPK(159–1191) or ANPK(159–789) variant that lacked most of the AR-interaction domain (Figure 6B). The activity of many Ser/Thr kinases and all known MAP kinases is regulated by either autophosphorylation or trans-phosphorylation within the activation segment in the catalytic subdomain VIII (Johnson et al., 1996). The putative activation segment of ANPK contains Ser357 and Tyr359 residues as potential autophosphorylation sites. Both residues were converted to Ala yielding the ANPK(S357A/Y359A) mutant. These substitutions attenuated ANPK kinase activity. However, ANPK(S357A/Y359A) was autophosphorylated to the same extent as full-length ANPK and ANPK(159–1191), whereas the C-terminally truncated form of the kinase, ANPK(159–789), exhibited no autophosphorylation (Figure 6C). The catalytic Asp322 in the subdomain VIB of ANPK is preceded by an Ala, and our data thus agree with the notion that phosphorylation is not required for activation of the kinases in which an Arg does not precede the conserved catalytic Asp (Johnson et al., 1996). Together, these results indicate that ANPK is not autophosphorylated at the putative activation segment and that autophosphorylation of this kinase is not a prerequisite for its catalytic activity in vitro.
Figure 6.
Protein kinase activity and autophosphorylation of different ANPK mutants. (A) Structural features of ANPK mutants studied. (B) Protein kinase activity of ANPK mutants. FLAG-tagged ANPK and indicated mutant proteins were expressed in CHO cells, immunoprecipitated using anti-FLAG monoclonal antibody, and subjected to immune complex kinase assays for 30 min at 30°C in the absence (–) and presence (+) of 5 μM MBP as described in MATERIALS AND METHODS. (C) Immune complex kinase assays of various ANPK forms in the absence of added substrate. Different ANPK forms were expressed to similar levels as judged by immunoblotting analysis (our unpublished data). (D) Phosphoamino acid analysis of autophosphorylated ANPK and MBP and c-Jun phosphorylated by ANPK in vitro. After in vitro kinase assay using immunopurified ANPK (1) and ANPK(S357A/Y359A) (2) from CHO cells, or MBP (3) and c-Jun (4) as substrates in the GST-ANPK(159–920) catalyzed phosphorylation, proteins were transferred onto PVDF membrane, radioactive bands corresponding to phosphorylated proteins were cut out and subjected to acid hydrolysis as described in MATERIALS AND METHODS. Phosphoamino acids were resolved by one-dimensional electrophoresis at pH 2.5. Positions of unlabeled internal phosphoamino acid standards are depicted.
Phosphoamino acid analysis of MBP and c-Jun phosphorylated by GST-ANPK(159–920) as well as that of autophosphorylated full-length ANPK and ANPK(S357A/Y359A), yielded only phosphoserine and phosphothreonine in each case, confirming that ANPK functions as a Ser/Thr kinase (Figure 6D).
ANPK Interacts with AR in Yeast and Mammalian Cells and under Cell-free Conditions
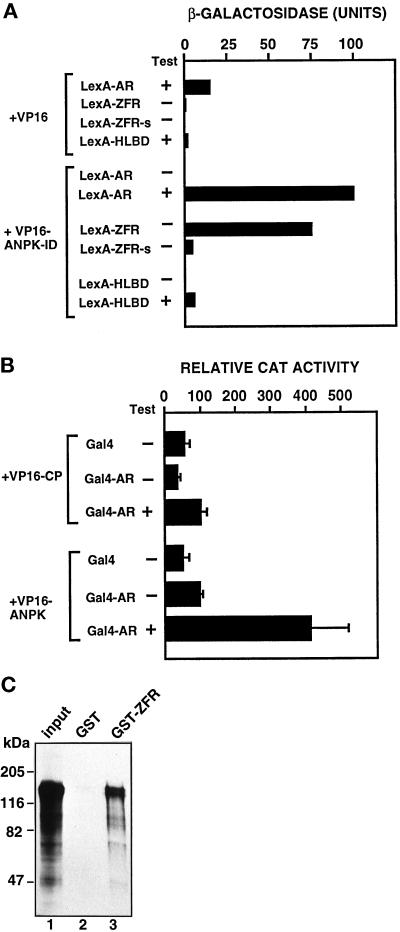
On the basis of sequences of the five ANPK clones obtained from the two-hybrid screen in yeast, residues 766–920 in the C-terminal region of ANPK appear to be sufficient for the interaction with AR. To characterize this interaction in more detail, full-length AR, AR ZFR without the hinge region residues 624–644 (ZFR-s), and a construct containing LBD and the hinge region (HLBD, residues 624–919 of hAR) were used to generate LexA-AR, LexA-ZFR-s, and LexA-HLBD fusion proteins. These proteins were expressed in yeast together with VP16 AD fused to amino acids 766–920 of ANPK (VP16-ANPK-ID). The interaction between full-length AR and VP16-ANPK-ID was androgen-dependent (Figure 7A). Unliganded AR may be unable to associate with ANPK due to steric hindrance presented by the hormone-free LBD or association of apo-AR with heat-shock proteins and/or other chaperones. Deletion of the hinge region residues from LexA-ZFR reduced markedly the interaction, and ANPK interacted only weakly with LexA-HLBD even in the presence of androgen (Figure 7A). Thus, in addition to the two zinc fingers, sequences in the hinge region are required for the interaction with ANPK.
Figure 7.
Interaction between AR and ANPK in yeast and mammalian cells and in vitro. (A) Plasmids expressing LexA or LexA fused to full-length AR (LexA-AR), AR ZFR including part of the hinge region (LexA-ZFR), AR ZFR without hinge region sequences (LexA-ZFR-s) or AR hinge-LBD fragment (LexA-HLBD) were introduced into L40 yeast cells together with expression plasmids for VP16 AD and VP16 AD fused to ANPK(766–920) (VP16-ANPK-ID). Transformants were grown in the presence (+) or absence (–) of 50 nM testosterone (Test). β-Gal activity in extracts from liquid yeast cultures are shown. Each bar depicts the average of three independent yeast transformants. Immunoblot and whole-cell ligand-binding assays indicated that the LexA-AR fusion proteins examined were expressed to comparable levels (our unpublished data). (B) Interaction of AR with ANPK in CV-1 cells. The ability of rAR and the DBD of Gal4 (Gal4-AR) as a fusion protein to interact with the residues 159–920 of ANPK fused to VP16 AD (VP16-ANPK) or with polyoma virus coat protein fused to VP16 AD (VP16-CP) was examined in CV-1 cells using the reporter plasmid pG5CAT. Cells (2.3 × 105 cells/35-mm dish) were transfected with 1.5 μg of each chimeric expression vector and 3 μg of pG5CAT reporter using DOTAP transfection reagent. Eighteen hours after transfection, the medium was changed to one containing charcoal-stripped 2% (vol/vol) FBS in the presence (+) or absence (–) of 100 nM testosterone (T), and the cells were incubated for an additional 30 h. Transcriptional activation is expressed as the relative CAT activity corrected for protein concentration. Mean ± SE values for at least three separate experiments are shown. (C) Specific interaction of ANPK and AR ZFR in vitro. 35S-Labeled full-length ANPK was synthesized by translation in vitro using reticulocyte lysate and incubated with GST alone (lane 2) or GST-AR ZFR (lane 3) adsorbed onto Glutathione Sepharose, after which the matrix was washed and bound proteins were analyzed as described in MATERIALS AND METHODS. Lane 1 represents 15% of the amount of labeled ANPK incubated with the matrix.
To determine whether AR interacts with ANPK also in mammalian cells, two-hybrid protein–protein interaction assays were carried out in CV-1 cells. Cotransfection of the cells with a Gal4-AR construct, full-length AR fused to the DNA-binding domain (DBD) of Gal4, and a plasmid encoding ANPK(159–920) fused to VP16 AD (VP16-ANPK) elicited a marked and androgen-dependent activation of the reporter gene, indicating hormone-enhanced recruitment of ANPK by AR (Figure 7B). When VP16 AD fused to polyoma virus coat protein (VP16-CP) was used instead of VP16-ANPK, only minimal reporter gene activity was observed in the presence of testosterone (Figure 7B).
Interaction of ANPK with AR under cell-free conditions was assessed by determining the ability of AR ZFR (residues 554–644) as a GST fusion protein to bind 35S-labeled ANPK(1–1191). A significant proportion of the input ANPK was retained on the GST-AR ZFR resin, whereas no binding on the control GST resin was observed (Figure 7C). Moreover, 35S-labeled luciferase did not adsorb onto GST-AR ZFR, verifying the specificity of the interaction between ANPK and AR ZFR (our unpublished data).
ANPK Colocalizes with AR in Mammalian Cells
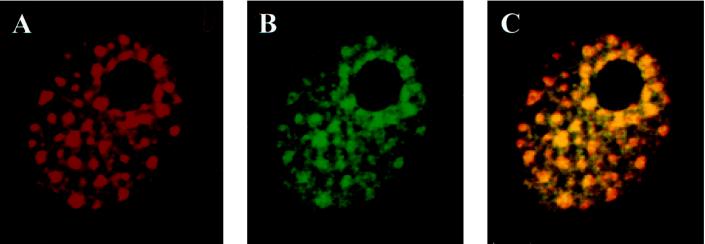
To assess whether ANPK and AR potentially interact in vivo, their colocalization was studied by a double immunofluorescence labeling technique followed by analysis with a confocal microscope. As already mentioned above, ANPK forms nuclear dot-like structures when transfected into CV-1 cells (Figure 3, A and B, and Figure 8A). We examined, therefore, whether cotransfection of AR with ANPK resulted in both proteins colocalizing to these subnuclear structures. As shown in Figure 8, subnuclear distribution of AR does exhibit the same pattern as that of ANPK. Superimposition of the images of ANPK and AR indicated that the majority of the two proteins had indeed colocalized. Transfection of AR alone did not show such a staining pattern: rather, it displayed a uniform nuclear distribution, suggesting that AR was recruited to nuclear dots by ANPK (our unpublished data). Our preliminary data also suggest that AR and ANPK colocalize in mouse mammary tumor S115 cells, which express relatively high levels of the two proteins.
Figure 8.
Colocalization of AR and ANPK in CV-1 cell nuclei. CV-1 cells seeded on glass cover slips on 10-cm plastic plates were cotransfected with 2.5 μg of FLAG-ANPK(2–1191) expression vector and 1 μg of rAR vector using DOTAP transfection reagent. Double immunofluorescence labeling was performed using anti-FLAG M2 monoclonal antibody and rabbit K183 antiserum for rAR as described in MATERIALS AND METHODS and analyzed by using a Bio-Rad MRC-1024 confocal laser scanning system connected to a Zeiss axiovert 100/135 microscope. Both ANPK (A, red) and AR (B, green) are present in nuclear dots, and the majority of the two proteins colocalize as revealed by the resulting yellow image following superimposition of A and B.
ANPK Enhances AR-dependent Transcriptional Activation
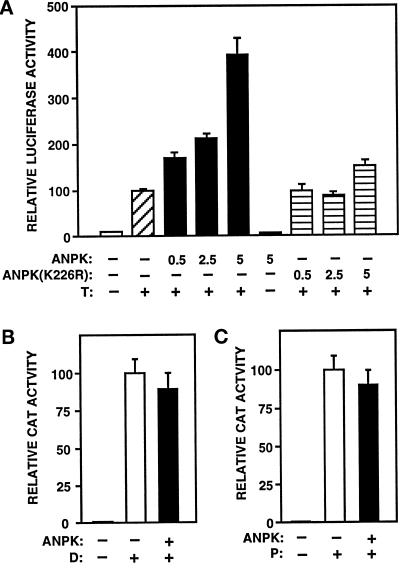
Transient transfection assays were used to examine consequences of ANPK overexpression on AR-dependent transcription. CV-1 cells were cotransfected with vectors encoding ANPK(159–1191) and rAR together with a reporter gene (luciferase) driven by the probasin promoter regulated by AR. Ligand-free rAR had minimal activity both in the absence and presence of ANPK (Figure 9A). The presence of androgen stimulated rAR activity ≥ 10-fold, and increasing amounts of coexpressed ANPK further activated AR function by three- to fourfold, whereas the kinase-negative ANPK(K226R) hardly influenced it (Figure 9A). The kinase-negative ANPK(K226R) inhibited the function of wild-type ANPK by 55 ± 6%, when the two proteins were coexpressed in a 3:1 ratio. ANPK did not modulate the antagonistic activity of the nonsteroidal antiandrogen casodex (our unpublished data). Very similar results were obtained in PC-3 and CHO cells. ANPK augmented AR-dependent transcription also from a minimal promoter containing two androgen response elements (AREs) in front of the E1b TATA sequence (pARE2-E1b-CAT) (see Figure 10B). It is of special interest that ANPK did not modulate the transcriptional activity of glucocorticoid receptor (GR) or progesterone receptor (PR) on pARE2-E1b-CAT expression under the same conditions (Figure 9, B and C).
Figure 9.
ANPK enhances androgen-induced transcriptional activation. (A) CV-1 cells were transfected using the calcium phosphate method with 5 μg of pPB(-285/+32)-LUC reporter plasmid along with 0.5 μg of pSG5-rAR and indicated amounts (μg) of pFLAG-ANPK(159–1191) or kinase-defective pFLAG-ANPK(K226R) in the presence or absence of 100 nM testosterone (T) as depicted. Total amount of DNA was kept constant by adding empty pFLAG-CMV-2 expression vector as needed. β-Gal expression plasmid, pCMVβ (2 μg/10-cm plate), was used to control for transfection efficiency. Luciferase (LUC) activities were normalized using β-gal activity. LUC activities are expressed relative to that of pSG5-rAR in the presence of testosterone (= 100), and the mean ± SE values of at least six independent experiments are given. (B and C) ANPK does not modulate PR- and GR-dependent transcription. (B) CV-1 cells were transfected with 5 μg of pARE2-E1b-CAT reporter containing two copies of the GRE/PRE/ARE motif of the rat tyrosine aminotransferase gene upstream of the adenovirus E1b TATA sequence along with 0.5 μg of pSG5-hGR, 5 μg of empty expression vector (pFLAG-CMV-2) (open bar) or pFLAG-ANPK(159–1191) (solid bar), and 2 μg of pCMVβ in the presence or absence of 100 nM dexamethasone (D). (C) CV-1 cells were transfected as in panel B, but using 0.5 μg of pSG5-hPR1 instead of pSG5-hGR in the presence or absence of 100 nM progesterone (P). CAT activities are normalized to β-gal activity and expressed relative to those achieved with pSG5-hGR or pSG5-hPR1 in the presence of P or D, respectively (= 100), and the mean ± SE values of at least three independent experiments are shown.
Figure 10.
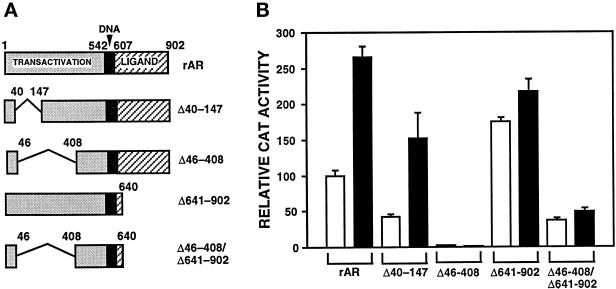
Influence of ANPK on the function of various AR mutants. (A) Structural features of AR mutants studied. (B) Effect of ANPK on AR mutants was examined in CV-1 cells by coexpressing rAR or the deletion mutants rAR▵40–147, rAR▵641–902, and rAR▵46–408/▵641–902 (0.5 μg of each pSG5 expression vector) in the presence of empty pFLAG-CMV2 expression vector (5 μg, open bars) or with pFLAG-ANPK(159–1191) (5 μg, solid bars) and 5 μg of pARE2-E1b-CAT reporter in the presence of 100 nM testosterone. Cells were transiently transfected using the calcium phosphate method. β-Gal expression plasmid, pCMVβ (2 μg/10-cm plate), was used to control for transfection efficiency. CAT activities are expressed relative to that of pSG5-rAR in the presence of testosterone (= 100), and the mean ± SE values of at least three independent experiments are given.
AF1 and LBD Are Also Required for ANPK to Activate AR Function
To identify the domains of AR that are critical for the activation by ANPK, various AR deletion mutants were investigated (Figure 10A). In addition to full-length AR, ANPK activated markedly the function of ARΔ40–147 lacking residues 40–147, whereas the function of two constitutively active AR forms, ARΔ641–902 and ARΔ46–408/Δ641–902, was barely influenced by coexpressed ANPK (Figure 10B). It is also noteworthy that ANPK did not activate a transcriptionally silent AR mutant, ARΔ46–408, devoid of activation function AF1 (Ikonen et al., 1997). These results indicate that the presence of both AF1 and LBD is required for ANPK to activate AR function.
ANPK Domains Needed to Activate AR Function
Several ANPK constructs (see Figure 6A) and AR were coexpressed in CV-1 cells, and their ability to activate AR-regulated probasin promoter was compared with that of ANPK(159–1191) (set as 100%). The activity of full-length ANPK(2–1191) did not differ markedly from that of ANPK(159–1191) (relative activity 82 ± 12% vs. 100 ± 9%). ANPK(159–772) that lacks most of the AR-interacting interface and ANPK(S357A/Y359A) exhibited both less than half of the activity of ANPK(159–1191) (35 ± 6% and 45 ± 9%, respectively). Diminished activity of ANPK(159–772) implies that the interaction between AR ZFR and ANPK is indeed important for ANPK to activate AR function. That ANPK(159–772) retained a modest activity on AR function can be explained by its remaining ability to phosphorylate some target substrates. Marked reduction in the activity of ANPK(S357A/Y359A) was not surprising, as the mutations in the putative activation segment also influenced its catalytic protein kinase activity.
Influence of ANPK on Cellular AR Concentration
To investigate whether ANPK influences the amount of AR, immunoreactive AR content and cellular androgen-binding capacity were analyzed in the presence and absence of cotransfected ANPK. There was a ∼2-fold increase in the amount of immunoreactive AR protein in samples derived from CV-1 cells expressing ANPK, whereas the amount of ARΔ641–902 devoid of LBD was not significantly influenced by coexpressed ANPK (Figure 11). Increased amount of immunoreactive wild-type AR antigen was consistent with ∼2.5-fold elevated androgen-binding capacity, as assessed by whole cell ligand-binding assays (AR + ANPK, 87, 520 ± 5470 receptors/cell vs. AR alone, 33,120 ± 2660 receptors/cell). ANPK did not increase the amount of AR mRNA and thus, it does not activate the promoters used to drive AR expression plasmids or stabilize AR mRNA. Together, these data suggest that ANPK–AR interaction stabilizes AR protein and, in addition to ZFR, LBD is needed for the stabilization to take place.
Figure 11.
Effect of ANPK on the amount of immunoreactive AR protein. CV-1 cells (2 × 106 cells/10-cm dish) were transfected by the calcium phosphate method with 5 μg of pSG5-rAR or pSG5-rAR▵641–902 together with 15 μg of pFLAG-CMV2, 15 μg of pFLAG-ANPK(2–1191) or pFLAG-ANPK(K226R) and cultured for 30 h in the presence of 100 nM testosterone. Each lane contained 20 μg of whole cell protein from cells transfected with pSG5-rAR plus pFLAG-CMV2 (lane 1), pSG5-rAR plus pFLAG-ANPK(2–1191) (lane 2), pSG5-rAR plus pFLAG-ANPK(K226R) (lane 3), pSG5-rAR▵641–902 plus pFLAG-CMV-2 (lane 4) and pSG5-rAR▵641–902 plus pFLAG-ANPK(2–1191) (lane 5). Immunoblot analysis was performed using rabbit K183 antiserum raised against full-length rAR (Karvonen et al., 1997) and immunoblots were developed using Amersham ECL western blotting detection reagents. Equal loading was confirmed by reprobing the membranes with an anti-α-tubulin antibody.
DISCUSSION
We have characterized in this work a novel Ser/Thr protein kinase, ANPK, which interacts with the AR. ANPK is a nuclear protein and widely expressed in human and rat tissues. The catalytic domain of ANPK is 39% identical with that of the rat minibrain gene product, a protein kinase whose Drosophila homologue is involved in postembryogenic neurogenesis (Tejedor et al., 1995). The human homologue of the minibrain gene was recently implicated in learning defects associated with Down syndrome (Smith et al., 1997). The kinase domain of ANPK also shows extensive similarity with Yak1 from Saccharomyces cerevisiae (Garrett and Broach, 1989). While this manuscript was being finalized for publication, the human counterpart of ANPK, termed putative protein kinase Y (PKY), was deposited to the GenBank (Begley et al., 1997). PKY comprises 1,215 residues and is 91% identical with ANPK. Expression of PKY mRNA was shown to be elevated in some multidrug-resistant cell lines, and PKY was postulated to play a role in the development of multidrug resistance (Begley et al., 1997).
The physiological substrates of Yak1 and MNB are not known. According to Hanks and Hunter (1995), Yak1 belongs to the Clk family in the CMGC group of protein kinases made up also by cdk, MAP kinase, and GSK3β/CK2 families. ANPK is likely to represent a new member of this protein kinase group. Our results show that the substrate specificity of ANPK is distinguishable from those of cdk and (ERK) MAP kinases. Extensive sequence similarity between ANPK and two cDNA fragments of homologous protein kinases also found in our two-hybrid screen (our unpublished results) suggests the existence of an additional subfamily of Ser/Thr protein kinases. Like MNB and Yak1, ANPK encompasses a distinct N-terminal domain, a kinase core domain and a characteristic C-terminal domain. Despite the similar molecular design of these kinases, the N-terminal and the long C-terminal domains of ANPK appear unique and share no significant homology with Yak1, MNB, or any other characterized protein kinase. Similar to the minibrain gene product (Song et al., 1997), both transiently expressed and endogenous ANPK protein resides in the nuclear compartment of cells.
The activity of transcription factors is frequently regulated by multiple phosphorylation and dephosphorylation events (Hunter and Karin, 1992; Hill and Treisman, 1995). Furthermore, steroid receptors are phosphoproteins, and many of them become hyperphosphorylated upon hormone binding, suggesting that there is a link between the phosphorylation status and the activation state of these receptors (Weigel, 1996). Activation of signal transduction cascades by kinase and phosphatase modulators can stimulate transcriptional activity of many steroid receptors, including AR, estrogen receptor (ER), GR, and PR (Beck et al., 1992; Somers and DeFranco, 1992; Cho and Katzenellenbogen, 1993; Moyer et al., 1993; Ikonen et al., 1994). Selective activation of extracellular signal-regulated kinases can also inhibit GR-mediated transcriptional activation (Rogatsky et al., 1998). In addition, various steroid receptors, such as human and rat ER, chicken PR, and human AR, may be activated in a ligand-independent manner by growth factors, neurotransmitters, and other compounds that modulate intracellular phosphorylation and/or dephosphorylation events (Denner et al., 1990; Power et al., 1991; Culig et al., 1994; Nazareth and Weigel, 1996). Mutagenesis experiments on the major phosphorylation sites of steroid receptors have, however, yielded contradictory results about the biological significance of receptor phosphorylation (Weigel, 1996).
Ligand-independent activation of chicken PR has been postulated to be mediated through changes in the phosphorylation of coregulatory proteins rather than in the receptor itself (Bai et al., 1997). By contrast, EGF signaling through the MAP kinase pathway induces phosphorylation of Ser118 in human ER, which results in the activation of this receptor in the absence of hormone (Kato et al., 1995; Bunone et al., 1996). A specific Tyr phosphorylation site is present within the carboxyl-terminal AF2 domain of ER (Arnold et al., 1997), and replacement of this residue with a charged amino acid or an Ala generated activated receptors that bound coactivator proteins in a ligand-independent manner (White et al., 1997). Collectively, these results emphasize that coregulatory proteins themselves and their interactions with steroid receptors are both likely to be regulated by protein phosphorylation events.
AR is phosphorylated on multiple sites, but nothing has been reported about the kinases involved (Zhou et al., 1995; Blok et al., 1998). Our current results indicate that both ERK2 and CDC2 are among the kinases that potentially regulate AR function through direct phosphorylation of the receptor protein. Interestingly, both of these kinases have recently been demonstrated to phosphorylate GR and thereby modulate its transcriptional activity (Krstic et al., 1997). Despite the physical interaction between ANPK and the receptor, AR was not phosphorylated by ANPK under cell-free conditions, and overexpression of the kinase did not increase the extent of AR phosphorylation in intact cells. Moreover, rARΔ40–147, devoid of the two potential phosphorylation sites of rAR (Ser61 and Ser75, corresponding to Ser81 and Ser94 residues of hAR) and a hAR mutant in which the third phosphorylation site, Ser650 (Zhou et al., 1995), was replaced by an Ala were both stimulated by ANPK in a manner similar to wild-type AR (Figure 10 and our unpublished results), supporting the notion that ANPK function does not involve direct phosphorylation of AR protein.
If ANPK does not indeed employ AR as the substrate, how might it then activate AR-dependent gene transcription? A modest increase in the receptor protein concentration in response to ANPK overexpression may not suffice. Anchoring of ANPK to AR would enable the kinase to recruit specific substrate proteins that are AR-associated proteins/coregulators, components of the transcriptional machinery, or chromatin structural proteins, such as histones. Phosphorylation of these proteins could subsequently enhance their interactions with the receptor or stabilize multiprotein complexes. While ANPK interacts with AR ZFR, the requirement of LBD for ANPK to activate AR-dependent transcription could be explained through a mechanism, in which ANPK phosphorylates a coactivator that interacts with LBD. Indeed, most of the recently characterized nuclear receptor coregulatory proteins seem to function through ligand-dependent interaction with the AF2 region in LBD (Horwitz et al., 1996). ANPK may also be involved in stabilization of the interaction between amino- and carboxyl-terminal regions of AR, an event known to be influenced by some other coregulatory proteins (Ikonen et al., 1997). ANPK showed strict substrate selectivity for nucleosomal core histones and catalyzed only phosphorylation of histone H3, a phenomenon that was recently suggested to be restricted to a small hyperacetylation-sensitive fraction of nucleosomes (Barratt et al., 1994). Therefore, it is tempting to speculate that ANPK is involved in the rearrangement of nucleosomes in promoter regions regulated by the AR.
ANPK did not activate PR and GR function under the experimental conditions used in this work, indicating that, even though many coregulators are shared by nuclear receptors (Horwitz et al., 1996), there is some receptor specificity among them. Action of cyclin D1 provides another example of receptor specificity: cyclin D1 is able to activate ER in a cdk-independent manner without exerting this action on other steroid receptors (Zwijsen et al., 1997). Moreover, activation differences between steroid receptors have been demonstrated in yeast cells expressing mutant forms of Hsp90 and Cdc37p (Bohen and Yamamoto, 1993; Fang et al., 1996; Fliss et al., 1997). Hsp90 was not phosphorylated by ANPK in vitro and thus not likely to play a role in ANPK-mediated activation of AR function (our unpublished observations).
Several nuclear receptor coregulators characterized over the last few years possess enzymatic activity, in that some catalyze acetylation of histones (Montminy, 1997; Perlmann and Evans, 1997) or even transcriptional activators (Gu and Roeder, 1997), whereas others are involved in deacetylation of histones (Pazin and Kadonaga, 1997; Perlmann and Evans, 1997). ANPK is a functionally active Ser/Thr kinase, and it appears to be the first putative steroid receptor coregulatory protein that catalyzes protein phosphorylation. However, to understand better the biological importance of ANPK, its physiological substrates should be identified and mechanisms controlling its catalytic activity clarified.
ACKNOWLEDGMENTS
The authors thank Ms. Leena Pietilä, Pirjo Kilpiö, and Seija Mäki for excellent technical assistance; Drs. P. Chambon, J.A. Cidlowski, M.G. Parker, and F.J. Rauscher III for plasmids; Dr. S.M. Hollenberg for providing the materials for the yeast two-hybrid system; and Dr. H. Vihinen for help in confocal microscopy. This work was supported by grants from the Medical Research Council of the Academy of Finland, the Finnish Foundation for Cancer Research, the Jalmari and Rauha Ahokas Foundation, the Research and Science Foundation of Farmos, and the University of Helsinki.
REFERENCES
- Adeyemo O, Kallio PJ, Palvimo JJ, Kontula K, Jänne OA. A single-base substitution in exon 6 of the androgen receptor gene causing complete androgen insensitivity: the mutated receptor fails to transactivate but binds to DNA in vitro. Hum Mol Genet. 1993;2:1809–1812. doi: 10.1093/hmg/2.11.1809. [DOI] [PubMed] [Google Scholar]
- Allgood VE, Oakley RH, Cidlowski JA. Modulation by vitamin B6 of glucocorticoid receptor-mediated gene expression requires transcription factors in addition to the glucocorticoid receptor. J Biol Chem. 1993;268:20870–20876. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan X-Y, Sauter G, Kallioniemi O-P, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Arnold SF, Melamed M, Vorojeikina DP, Notides AC, Sasson S. Estradiol-binding mechanism and binding capacity of the human receptor is regulated by tyrosine phosphorylation. Mol Endocrinol. 1997;11:48–53. doi: 10.1210/mend.11.1.9876. [DOI] [PubMed] [Google Scholar]
- Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Bai W, Rowan BG, Allgood VE, O’Malley BW, Weigel NL. Differential phosphorylation of chicken progesterone receptor in hormone-dependent and ligand-independent activation. J Biol Chem. 1997;272:10457–10463. doi: 10.1074/jbc.272.16.10457. [DOI] [PubMed] [Google Scholar]
- Barratt MJ, Hazzalin CA, Cano E, Mahadevan LC. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc Natl Acad Sci USA. 1994;91:4781–4785. doi: 10.1073/pnas.91.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Chien CT, Sternglanz R, Fields S. Using the two-hybrid system to detect protein–protein interactions. In: Hartley DA, editor. Cellular Interactions in Development: A Practical Approach. Oxford, UK: IRL; 1993. pp. 153–179. [Google Scholar]
- Begley DA, Berkenpas MB, Sampson KE, Abraham I. Identification and sequence of human PKY, a putative kinase with increased expression in multidrug-resistant cells, with homology to yeast protein kinase Yak1. Gene. 1997;200:35–43. doi: 10.1016/s0378-1119(97)00350-8. [DOI] [PubMed] [Google Scholar]
- Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Beato M, Sanchez-Pacheco A. Interaction of steroid hormone receptors with the transcription initiation complex. Endocr Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- Beck CA, Weigel NL, Edwards DP. Effects of hormone and cellular modulators of protein phosphorylation on transcriptional activity, DNA binding, and phosphorylation of human progesterone receptors. Mol Endocrinol. 1992;6:607–620. doi: 10.1210/mend.6.4.1316549. [DOI] [PubMed] [Google Scholar]
- Blanco JCG, Wang I-M, Tsai SY, Tsai M-J, O’Malley BW, Jurutka PW, Haussler MR, Ozato K. Transcription factor TFIIB and vitamin D receptor cooperatively activate ligand-dependent transcription. Proc Natl Acad Sci USA. 1995;92:1535–1539. doi: 10.1073/pnas.92.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok LJ, de Ruiter PE, Brinkmann AO. Foskolin-induced dephosphorylation of the androgen receptor impairs ligand binding. Biochemistry. 1998;37:3850–3857. doi: 10.1021/bi9724422. [DOI] [PubMed] [Google Scholar]
- Bohen SP, Yamamoto KR. Modulation of steroid receptor signal transduction by heat shock protein. In: Morimoto R, Tissieres A, Georgopoulos C, editors. The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 313–334. [Google Scholar]
- Bunone G, Briand P-A, Miksicek RJ, Picard D. Activation of the unliganded estrogen receptor involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- Cavaillés V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Juguilon H, Montminy M, Evans RM. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Cho H, Katzenellenbogen BS. Synergistic activation of estrogen receptor-mediated transcription by estradiol and protein kinase activators. Mol Endocrinol. 1993;7:441–452. doi: 10.1210/mend.7.3.7683375. [DOI] [PubMed] [Google Scholar]
- Culig Z, Hobish A, Cronauer M, Radmayr C, Trapman J, Hittmair A, Bartsch G, Klocker H. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res. 1994;54:5474–5478. [PubMed] [Google Scholar]
- Denner LA, Weigel NL, Maxwell BL, Schrader WT, O’Malley BW. Regulation of progesterone receptor-mediated transcription by phosphorylation. Science. 1990;250:1740–1743. doi: 10.1126/science.2176746. [DOI] [PubMed] [Google Scholar]
- Eastman A. An improvement to the novel rapid assay for chloramphenicol acetyltransferase gene expression. Biotechniques. 1987;5:73. [Google Scholar]
- Fang Y, Fliss AE, Robins DM, Caplan AJ. Hsp90 regulates androgen receptor hormone binding affinity in vivo. J Biol Chem. 1996;271:28697–28702. doi: 10.1074/jbc.271.45.28697. [DOI] [PubMed] [Google Scholar]
- Fliss AE, Fang Y, Boschelli F, Caplan AJ. Differential in vivo regulation of steroid hormone receptor by Cdc37p. Mol Biol Cell. 1997;8:2501–2509. doi: 10.1091/mbc.8.12.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Broach JR. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAK1, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol Cell Biol. 1991;11:4045–4052. doi: 10.1128/mcb.11.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hadzig E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka BM, Samuels HH. A 10-amino acid sequence in the N-terminal A/B domain of thyroid hormone receptor a is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. The eucaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM. The protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz KB, Jackson TA, Bain DL, Richard JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- Hunter T, Karin M. Regulation of transcription by phosphorylation. Cell. 1992;70:375–385. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ, Jänne OA. Interaction between amino- and carboxyl-terminal regions of rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ, Kallio PJ, Reinikainen P, Jänne OA. Stimulation of androgen-regulated transactivation by modulators of protein phosphorylation. Endocrinology. 1994;135:1359–1366. doi: 10.1210/endo.135.4.7925097. [DOI] [PubMed] [Google Scholar]
- Ing NH, Beekman JM, Tsai SY, Tsai M-J, O’Malley BW. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- Jelinek T, Weber MJ. Optimization of the resolution of phosphoamino acids by one-dimensional thin-layer electrophoresis. Biotechniques. 1993;15:629–630. [PubMed] [Google Scholar]
- Johnson LN, Noble MEM, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Kallio PJ, Poukka H, Moilanen A, Jänne OA, Palvimo JJ. Androgen receptor mediated transcriptional regulation in the absence of direct interaction with a specific DNA element. Mol Endocrinol. 1995;9:1017–1028. doi: 10.1210/mend.9.8.7476976. [DOI] [PubMed] [Google Scholar]
- Kamei Y, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Karvonen U, Kallio PJ, Jänne OA, Palvimo JJ. Interaction of androgen receptors with androgen response elements in intact cells: roles of amino- and carboxyl-terminal regions and the ligand. J Biol Chem. 1997;272:15973–15979. doi: 10.1074/jbc.272.25.15973. [DOI] [PubMed] [Google Scholar]
- Kato S, et al. Activation of estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Kentrup H, Becker W, Heukelbach J, Wilmes A, Schürmann A, Huppertz C, Kainulainen H, Joost H-G. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J Biol Chem. 1996;271:3488–3495. doi: 10.1074/jbc.271.7.3488. [DOI] [PubMed] [Google Scholar]
- Krstic MD, Rogatsky I, Yamamoto KR, Garabedian MJ. Mitogen-activated and cyclin-dependent protein kinases selectively and differentially modulate transcriptional enhancement by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3947–3954. doi: 10.1128/mcb.17.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;22:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the liganddependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Ryan F, Swaffield SA, Johnston SA, Moore DD. Interaction of thyroid hormone receptor with a conserved transcriptional mediator. Nature. 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- Lichtarge O, Yamamoto KR, Cohen FE. Identification of functional surfaces of the zinc binding domains of intracellular receptors. J Mol Biol. 1997;274:325–337. doi: 10.1006/jmbi.1997.1395. [DOI] [PubMed] [Google Scholar]
- McEwan IJ, Gustafsson J-Å. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci USA. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengus G, May M, Carré L, Chambon P, Davidson I. Human TAFII135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- Moilanen, A.-M., Poukka, H., Karvonen, U., Häkli, M., Jänne, O.A., and Palvimo, J.J. (1998). Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Moilanen A-M, Rouleau N, Ikonen T, Palvimo JJ, Jänne OA. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 1997;412:355–358. doi: 10.1016/s0014-5793(97)00791-6. [DOI] [PubMed] [Google Scholar]
- Montminy M. Something new to hang your HAT on. Nature. 1997;387:654–655. doi: 10.1038/42594. [DOI] [PubMed] [Google Scholar]
- Moyer ML, Borror KC, Bona BJ, DeFranco DB, Nordeen SK. Modulation of cell signalling pathways can enhance or impair glucocorticoid-induced gene expression without altering the state of receptor phosphorylation. J Biol Chem. 1993;268:22933–22940. [PubMed] [Google Scholar]
- Nazareth VL, Weigel NL. Activation of the human androgen receptor through a protein kinase A signalling pathway. J Biol Chem. 1996;271:19900–19907. doi: 10.1074/jbc.271.33.19900. [DOI] [PubMed] [Google Scholar]
- Oñate SA, Tsai SY, Tsai M-J, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Palvimo JJ, Kallio PJ, Ikonen T, Mehto M, Jänne OA. Dominant negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol Endocrinol. 1993;7:1399–1028. doi: 10.1210/mend.7.11.8114755. [DOI] [PubMed] [Google Scholar]
- Palvimo J, Mäenpää PH. Binding of high-mobility-group proteins HMG 14 and HMG 17 to DNA and histone H1 as influenced by phosphorylation. Biochim Biophys Acta. 1988;952:172–180. doi: 10.1016/0167-4838(88)90113-6. [DOI] [PubMed] [Google Scholar]
- Palvimo JJ, Partanen M, Jänne OA. Characterization of a cell specific modulatory element in the murine ornithine decarboxylase promoter. Biochem J. 1996;316:993–998. doi: 10.1042/bj3160993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palvimo JJ, Reinikainen P, Ikonen T, Kallio PJ, Moilanen A, Jänne OA. Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Evans RM. Nuclear receptors in Sicily: all in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- Power RF, Mani SK, Codina J, Conneely OM, O’Malley BW. Dopaminergic and ligand-independent activation of steroid hormone receptors. Science. 1991;254:1636–1639. doi: 10.1126/science.1749936. [DOI] [PubMed] [Google Scholar]
- Quigley CA, DeBellis A, Marschke KB, El-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Logan SK, Garabedian MJ. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc Natl Acad Sci USA. 1998;95:2050–2055. doi: 10.1073/pnas.95.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- Schena M, Freedman LP, Yamamoto KR. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989;3:1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- Schulman IG, Chakravarti D, Juguilon H, Romo A, Evans RM. Interactions between the retinoid X receptor and a conserved region of the TATA-binding protein mediate hormone-dependent transactivation. Proc Natl Acad Sci USA. 1995;92:8288–8292. doi: 10.1073/pnas.92.18.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Klotzbucher M, Sachs M, Uber V, Klein-Hitpass L. Identification of a transactivation function in the progesterone receptor that interacts with the TAFII110 subunit of the TFIID complex. J Biol Chem. 1995;270:21331–21338. doi: 10.1074/jbc.270.36.21331. [DOI] [PubMed] [Google Scholar]
- Shindoh N, Kudoh J, Hideto M, Yamaki A, Minoshima M, Shimizu Y, Shimizu N. Cloning of a human homolog of the Drosophila Minibrain/rat Dyrk gene from “the Down syndrome critical region” of chromosome 21. Biochem Biophys Res Commun. 1996;225:92–99. doi: 10.1006/bbrc.1996.1135. [DOI] [PubMed] [Google Scholar]
- Smith DJ, et al. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat Genet. 1997;16:28–36. doi: 10.1038/ng0597-28. [DOI] [PubMed] [Google Scholar]
- Somers JP, DeFranco DB. Effects of okadaic acid, a protein phosphatase inhibitor, on glucocorticoid receptor mediated enhancement. Mol Endocrinol. 1992;6:26–34. doi: 10.1210/mend.6.1.1310797. [DOI] [PubMed] [Google Scholar]
- Song W-J, Chung S-H, Kurnit DM. The murine Dyrk protein maps to chromosome 16, localizes to the nucleus, and can form multimers. Biochem Biophys Res Commun. 1997;231:640–644. doi: 10.1006/bbrc.1997.6154. [DOI] [PubMed] [Google Scholar]
- Starr DB, Matsui W, Thomas JR, Yamamoto KR. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- Tejedor F, Zhu XR, Kaltenbach E, Ackerman A, Bauman A, Canal I, Heisenberg M, Fischbach KF, Pongs O. Minibrain: a new protein kinase family involved in postembryogenetic neurogenesis in Drosophila. Neuron. 1995;14:287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Hiene MJS, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- vom Baur E, Zechel C, Heery D, Heine MJS, Garnier JM, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- Weigel NL. Steroid hormone receptors and their regulation by phosphorylation. Biochem J. 1996;319:657–667. doi: 10.1042/bj3190657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Sjöberg M, Kalkhoven E, Parker MG. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z-X, Lane VM, Kemppainen JA, Wilson EM. Identification of three proline-directed phosphorylation sites in the human androgen receptor. Mol Endocrinol. 1995;9:605–615. doi: 10.1210/mend.9.5.7565807. [DOI] [PubMed] [Google Scholar]
- Zwijsen RML, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJAM. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]