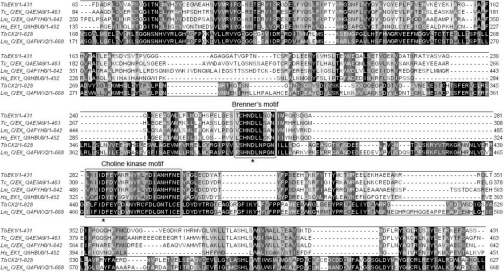
Figure 2. A Tcoffee alignment of the predicted amino acid sequences of TbEK1 (formerly TbC/EK1) and TbC/EK2 with CKs and EKs of other eukaryotic organisms: T. cruzi (Swiss-Prot accession number Q4E3A9), L. major (Q4FYH6 and Q4FWX2) and Homo sapiens (Q9HBU6).
Residues are shaded according to percentage similarity using the Jalview 2.08.1 software. The boxes highlight the choline kinase motif and Brenner's phosphotransferase motif. The two aspartate residues marked by an asterisk were mutated in TbEK1 to generate the catalytically inactive mutants D267A and D286A.