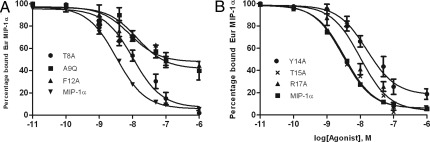
Fig. 4.
Binding of MIP-1α variants to CCR5. Competition binding curves were generated on membranes from CHO cells expressing CCR5 and using 8 nM Eur-MIP-1α as a trace. Example inhibition data are shown for T8A, A9Q, and F12A (A), and Y14A, T15A, and R17A, (B) and all results are summarized in Table 1. The data were normalized for nonspecific binding and maximal specific binding in the absence of competitor (100%). The results were analyzed with GraphPad PRISM software by using a single-site model. All points were run in triplicate (error bars indicate SEM), and the presented data are a representative of two independent experiments, each done in duplicate. See Fig. S2 for binding curves for other mutants.