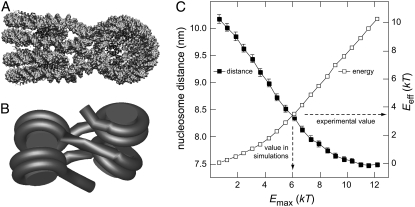
FIGURE 3.
Calibration of internucleosomal attraction energies. (A) Crystal structure of the tetranucleosome particle (23). Some bending of the linker DNA is induced to promote nucleosome-nucleosome stacking. (B) Coarse-grained model of the tetranucleosome used in the MC simulations. (C) Plot of mean nucleosome-nucleosome distance (▪) and mean effective internucleosomal attraction energy Eeff (□) versus the input value Emax. The parameter Emax is defined as the nucleosome-nucleosome interaction energy obtained for an optimal stacking of two nucleosomes. Data points represent averages of ∼1000 independent configurations with the error bars indicating the 95% confidence interval. An experimental value of Eeff = 3.4 kT was derived from chromatin stretching experiments (42). In the simulation, choosing Emax = 6.1 kT resulted in an effective interaction strength of Eeff = 3.5 kT. These values were used for all MC simulations of this study.