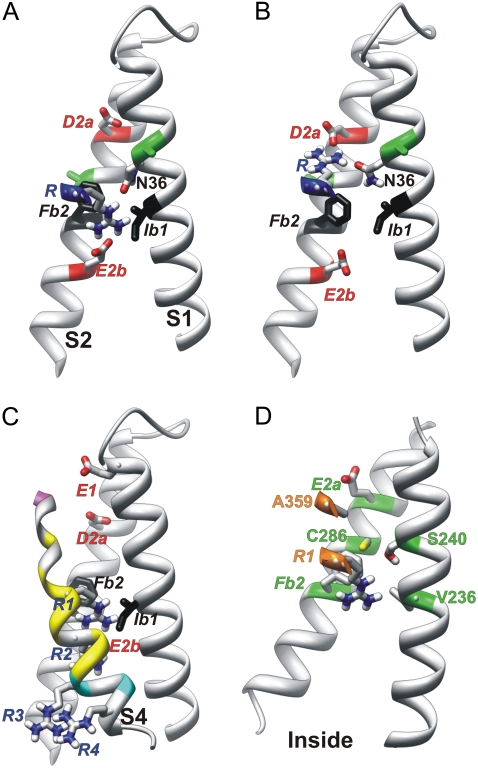
FIGURE 4.
Side view of S1, S2, and portions of S4 of Closedb models. Side chains of N36, D2a, E2b, and an S4 arginine are colored by atomic element (red, oxygen; blue, nitrogen; white, hydrogen; gray, carbon; and yellow, sulfur). Transition barrier residues are black. (A and B) An S4 arginine is oriented (A) inwardly for negative voltages, where it salt-bridges to the E2b group, and (B) outwardly for positive voltages, where it salt-bridges to the D2a group. Although this representation was generated from Closed2b and Closed3b models in which the arginine is R3, it could be used for Closedb models favored at more negative voltages, in which the S4 arginine would be R1 or R2. Green positions on S1 and S2 (A and B) indicate residues analogous to Shaker I241C and I287C, which can cross-link with the R1C residue at negative voltages in Shaker channels (24). (C) Closed0b model, in which S4 residues are colored according to accessibility to [2-(trimethylammonium)ethyl]-methanethiosulfonatein at negative voltages of introduced cysteines at analogous positions in Shaker channels (25), using alignment 2. Magenta indicates accessibility from the outside, cyan accessibility from the inside, and yellow inaccessibility from either side. (D) Closed0b model of the Shaker channel illustrates residue positions where mutations have a strong, apparently steric effect on Ω currents (27).