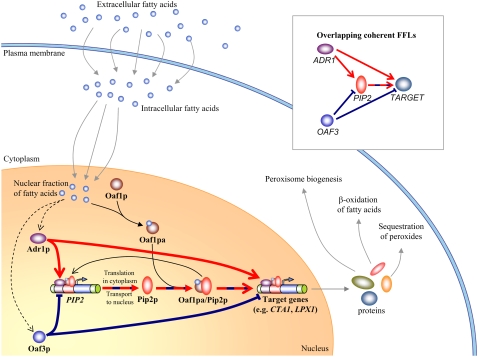
FIGURE 1.
The yeast oleate-responsive transcriptional network contains an OFFNM. Lines terminating in open arrows and blunted lines represent transcriptional up- and down-regulation, respectively. Lines terminating in solid arrows indicate molecular processes such as transport, transcription/translation, and dimerization. Dotted black arrows indicate indirect carbon-source-dependent activation. Red and blue arrows and blunted lines represent the Adr1p- and Oaf3p-driven coherent feed-forward motifs, respectively. The alternating red/blue dashed line represents the overlapping region. The inset panel demonstrates schematically the OFFNM. Intracellular FA (oleate) binds Oaf1p, activating the TF. Active Oaf1p forms a heterodimer with Pip2p, and this heterodimer targets the ORE on DNA as a transcriptional activator. The promoter of the gene PIP2 contains an ORE, and thus PIP2 is transcriptionally autoregulated in the presence of oleate. Adr1p is rapidly activated in the presence of nonfermentable carbon sources and targets UAS1 elements in the promoters of the target genes PIP2 and CTA1. Adr1p therefore drives a coherent feed-forward network motif targeting PIP2 and CTA1 (thick red lines). Oaf3p is a transcriptional inhibitor whose target footprint (in terms of number of genes) is strongly increased under oleate growth conditions (17). It drives a coherent inhibitory feed-forward network motif targeting PIP2 and CTA1 (thick blue lines).