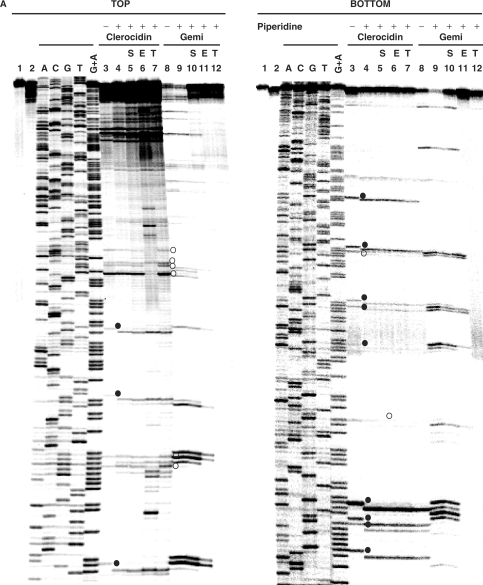
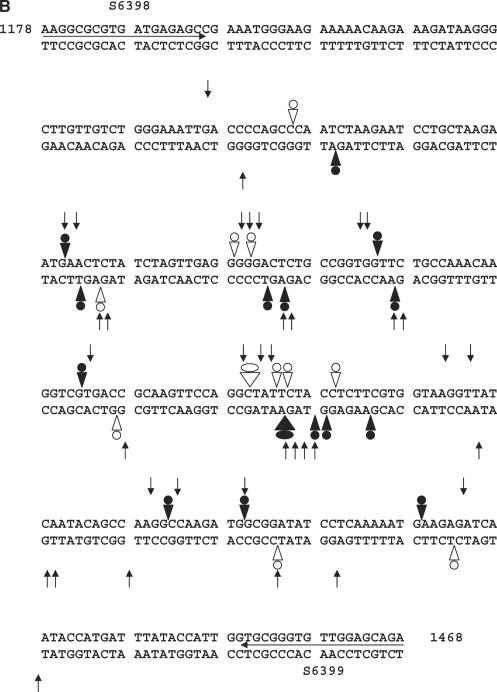
Figure 4.
CL induces DNA modification and irreversible/reversible DNA breakage at gyrase cleavage sites. (A) Probing CL modification and cleavage reversibility. The S fragment, 33P-labelled at the 5′ end of the TOP or BOTTOM strand, was incubated with S. pneumoniae gyrase and 1 mM ATP (lanes 2–12) in the absence (lane 2) or presence of 200 μM CL (lanes 3–7) or 100 μM gemifloxacin (lanes 8–12). Samples in lanes 5 and 10, 6 and 11 and 7 and 12 were further incubated with 0.6 M NaCl for 15 min (denoted by S), with 35 mM EDTA for 15 min (E), or heating to 65°C for 15 min (T), respectively. After cleavage induction with SDS and proteinase K, DNA was precipitated with ethanol. Cleavage products in lanes 4–7 and 9–12 were further incubated with 1-M piperidine at 90°C for 30 min and then lyophilized. Reaction products were loaded and run on 6% denaturing polyacrylamide gels alongside the G + A Maxam–Gilbert chemical sequencing products, and the ACGT dideoxy sequencing products obtained using the same 5′ end-labelled oligonucleotide primers. Lane 1 is untreated substrate DNA. Lane 2 is a control for gyrase-processed DNA. Lanes 3 and 8 are cleavage products that were not treated with piperidine. DNA cleavage products that undergo shortening after hot piperidine are denoted by filled circles; those unaffected by piperidine are labelled with open circles. Gemifloxacin cleavage products were uniformly unaffected by hot piperidine (compare lanes 8 and 9) and were formed reversibly (lanes 10–12). Several new bands appearing after EDTA treatment (TOP strand, lane 6) may arise from sealing of sites proximal to the 5′ end revealing distal cleavage sites. (B) Mapping CL-gyrase cleavage sites in the S fragment. Arrowheads and arrows show cleavage sites stimulated by CL and gemifloxacin, respectively. Filled and open arrowheads denote sites of irreversible and reversible cleavage, respectively. Filled and open circles denote CL sites at which the 3′ nucleotide was released by hot piperidine or was wholly (or partially) unaffected by piperidine treatment, respectively. The large arrowheads and circles denote the most efficient cleavage site. Different to the 4-bp gyrase stagger, two sites with an apparent 2-bp overhang likely arise from closely spaced single cuts. Numbers refer to nucleotide positions in the S. pneumoniae parE gene (31) counting from the first nucleotide of the initiation codon ATG. Horizontal arrows denote the sequences of forward (S6398) and reverse oligonucleotides (S6399) used in PCR.