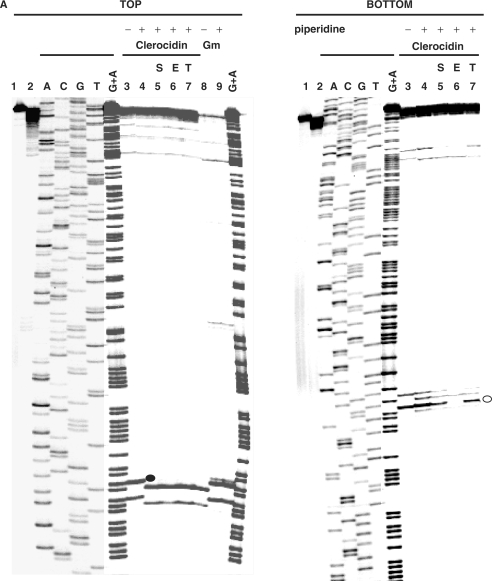
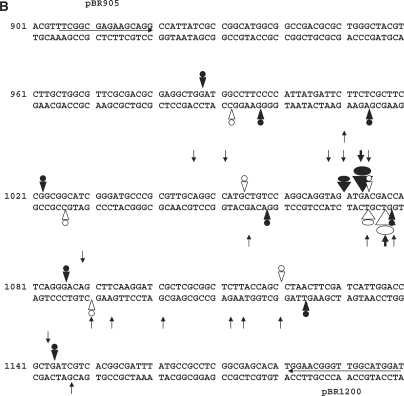
Figure 5.
CL-stimulated gyrase cleavage at the pBR322 ‘1073’ site. (A) DNA modification and EDTA reversal. A 234-bp pBR322 fragment (nucleotides 966–1200), labelled with 33P at the 5′ end of the TOP or BOTTOM strand, was incubated with gyrase and 1 mM ATP in the absence (lane 2) or presence of 200 μM CL (lanes 3–7) or 100 μM gemifloxacin (Gm) (lanes 8 and 9). Samples in lanes 5–7 were further incubated with NaCl (S), EDTA (E) or at 65°C (T) as described in the Figure 4 legend. After treatment with SDS and proteinase K, reaction products were precipitated with ethanol. Samples in lanes 4–7 and 9 were incubated with hot piperidine. Samples were lyophilized and run on 6% denaturing polyacrylamide gels alongside G+A chemical sequencing and ACGT chain termination sequencing products. Lanes 1 and 2 are controls for untreated DNA and gyrase processed DNA, respectively. Lanes 3 and 8 are non-piperidine treated cleavage products. The main cleavage site at 1073 produced CL-stabilized fragments indicated by a filled circle (TOP strand, piperidine-shifted and irreversible) and by an unfilled circle (BOTTOM strand, largely unshifted by piperidine and EDTA reversible). (B) Location of DNA gyrase cleavage sites in the pBR322 fragment. Symbols are used as described in the Figure 4B legend. Large arrowheads denote the ‘1073’ site; smaller arrowheads indicate weaker sites. Nucleotide sequence from ref 30.