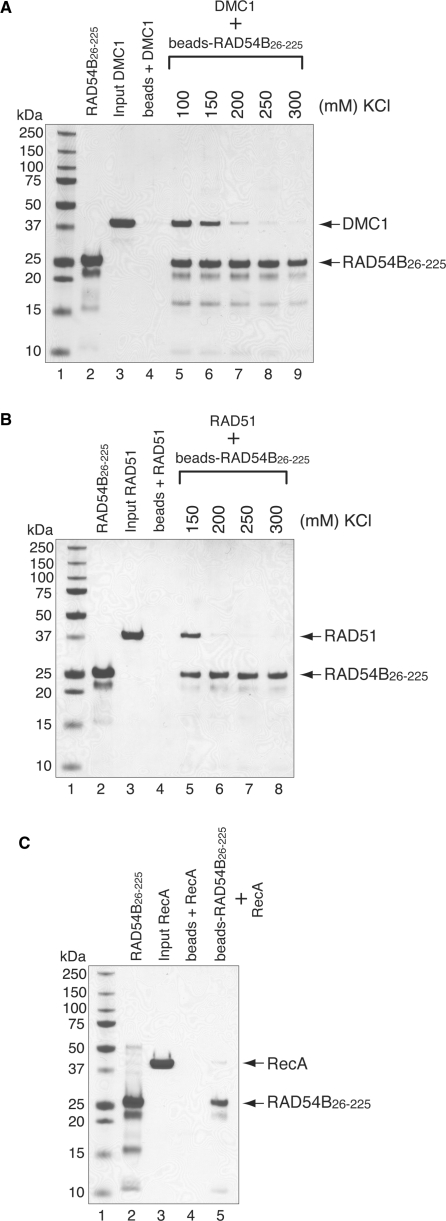
Figure 3.
RAD54B26–225 interacts with RAD51 and DMC1. The interactions were observed by a pull-down assay, in which DMC1 (A) or RAD51 (B) was mixed with RAD54B26–225 that was covalently conjugated to an Affi-Gel 15 matrix. The proteins bound to the RAD54B26–225-conjugated beads were eluted by SDS–PAGE sample buffer, and fractionated on a 12% SDS–PAGE gel. Lanes 2 and 3 are one-tenth of the total proteins used. Lane 4 is the negative control using the Affi-Gel 15 matrix without RAD54B26–225. The salt concentration was titrated for both binding experiments, which are shown beyond lane 5. (C) Interaction between bacterial RecA and RAD54B26–225. The binding experiment was performed in the presence of 100 mM KCl. The bands were visualized by Coomassie Brilliant Blue staining.