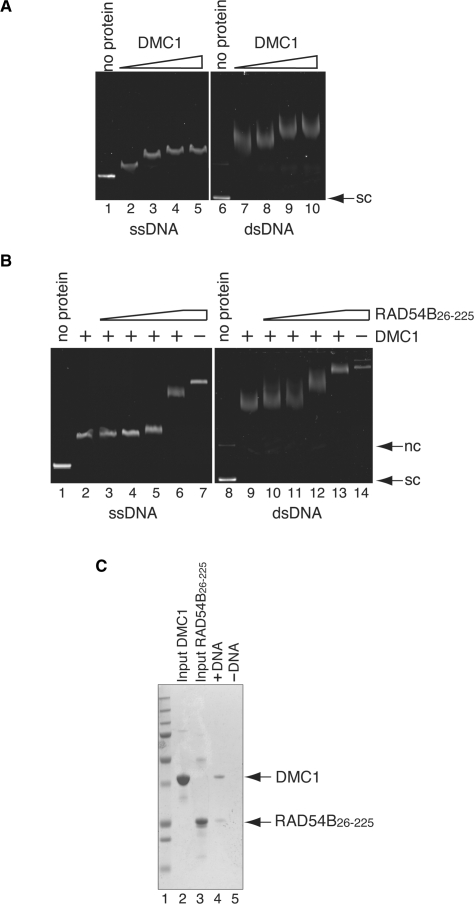
Figure 4.
Interaction between DMC1 and RAD54B26–225 on DNA. (A) DNA-binding activity of DMC1. Increasing amounts of DMC1 (10, 20, 40 and 80 μM in lanes 2–5 and lanes 7–10, respectively) were incubated with φX174 ssDNA (20 μM in nucleotides) or φX174 superhelical dsDNA (10 μM in nucleotides). Lanes 1 and 6 indicate negative control experiments without protein. The reaction mixtures were fractionated on a 1% agarose gel, which was stained with ethidium bromide. (B) RAD54B26–225 forms ternary complexes with DMC1 and DNA. A constant amount of DMC1 (40 μM) was incubated with φX174 circular ssDNA (20 μM in nucleotides) or φX174 superhelical dsDNA (10 μM in nucleotides) at 37°C for 20min, followed by an incubation with increasing amounts of RAD54B26–225 (0, 2.0, 4.0, 8.0 and 16 μM in lanes 2–6 and lanes 9–13, respectively) at 37°C for 20 min. In lanes 7 and 14, RAD54B26–225 (16 μM) was incubated with ssDNA and dsDNA, but not DMC1, respectively. Lanes 1 and 8 indicate negative control experiments without protein. (C) Electroelution analysis of the protein–DNA complex. The protein–DNA complex detected in Figure 4B lane 13 was electroeluted from the agarose gel, and was analyzed by 12% SDS–PAGE gel (lane3). Lane 5 is the negative control experiment performed without dsDNA. Lane 1 indicates the molecular mass markers. Lanes 2 and 3 are one-tenth of the input DMC1 and RAD54B26–225, respectively. Nc and sc indicate nicked circular and superhelical dsDNA, respectively.